3BD1
 
 | | Structure of the Cro protein from putative prophage element Xfaso 1 in Xylella fastidiosa strain Ann-1 | | Descriptor: | CHLORIDE ION, Cro protein, GLYCEROL, ... | | Authors: | Hall, B.M, Roberts, S.A, Montfort, W.R, Cordes, M.H. | | Deposit date: | 2007-11-13 | | Release date: | 2008-03-25 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Transitive homology-guided structural studies lead to discovery of
Cro proteins with 40% sequence identity but different folds
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
6ON0
 
 | | STRUCTURE OF N15 CRO COMPLEXED WITH CONSENSUS OPERATOR DNA | | Descriptor: | DNA (5'-D(*TP*TP*TP*AP*TP*AP*GP*CP*TP*AP*GP*CP*TP*AP*TP*AP*A)-3'), Gp39 | | Authors: | Hall, B.M, Roberts, S.A, Cordes, M.H.J. | | Deposit date: | 2019-04-19 | | Release date: | 2019-05-15 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Extreme divergence between one-to-one orthologs: the structure of N15 Cro bound to operator DNA and its relationship to the lambda Cro complex.
Nucleic Acids Res., 47, 2019
|
|
2OVG
 
 | | Lambda Cro Q27P/A29S/K32Q triple mutant at 1.35 A in space group P3221 | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Phage lambda Cro, SULFATE ION | | Authors: | Hall, B.M, Heroux, A, Roberts, S.A, Cordes, M.H. | | Deposit date: | 2007-02-13 | | Release date: | 2008-01-08 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Two structures of a lambda Cro variant highlight dimer flexibility but disfavor major dimer distortions upon specific binding of cognate DNA.
J.Mol.Biol., 375, 2008
|
|
2ECS
 
 | | Lambda Cro mutant Q27P/A29S/K32Q at 1.4 A in space group C2 | | Descriptor: | ACETATE ION, CHLORIDE ION, LITHIUM ION, ... | | Authors: | Hall, B.M, Roberts, S.A, Cordes, M.H. | | Deposit date: | 2007-02-14 | | Release date: | 2008-01-08 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Two structures of a lambda Cro variant highlight dimer flexibility but disfavor major dimer distortions upon specific binding of cognate DNA.
J.Mol.Biol., 375, 2008
|
|
7M1L
 
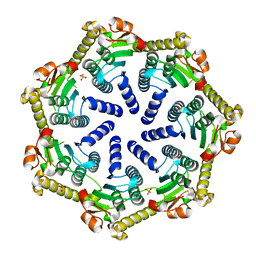 | | Crystal structure of Pseudomonas aeruginosa ClpP2 | | Descriptor: | ATP-dependent Clp protease proteolytic subunit, PHOSPHATE ION | | Authors: | Hall, B.M, Grant, R.A, Baker, T.A, Sauer, R.T. | | Deposit date: | 2021-03-13 | | Release date: | 2021-03-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | ClpP1P2 peptidase activity promotes biofilm formation in Pseudomonas aeruginosa.
Mol.Microbiol., 115, 2021
|
|
7A29
 
 | |
6ZXN
 
 | |
7A25
 
 | |
3BXD
 
 | | Crystal structure of Mouse Myo-inositol oxygenase (re-refined) | | Descriptor: | 1,2,3,4,5,6-HEXAHYDROXY-CYCLOHEXANE, FE (III) ION, FORMIC ACID, ... | | Authors: | Hallberg, B.M. | | Deposit date: | 2008-01-13 | | Release date: | 2008-02-05 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and biophysical characterization of human myo-inositol oxygenase.
J.Biol.Chem., 283, 2008
|
|
2FIM
 
 | | Structure of the C-terminal domain of Human Tubby-like protein 1 | | Descriptor: | 3-(N,N-DIMETHYLOCTYLAMMONIO)PROPANESULFONATE, SULFATE ION, Tubby related protein 1 | | Authors: | Hallberg, B.M, Ogg, D, Arrowsmith, C, Berglund, H, Edwards, A, Ehn, M, Flodin, S, Graslund, S, Hammarstrom, M, Hogbom, M, Holmberg-Schiavone, L, Kotenyova, T, Kursula, P, Nilsson-Ehle, P, Nordlund, P, Nyman, T, Sagemark, J, Stenmark, P, Sundstrom, M, Thorsell, A.G, Van Den Berg, S, Weigelt, J, Persson, C. | | Deposit date: | 2005-12-29 | | Release date: | 2006-02-07 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of the C-terminal domain of Human Tubby-like protein 1
To be published
|
|
8A99
 
 | | SARS Cov2 Spike in 1-up conformation complex with Fab47 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Fab47 (Heavy chain variable domain), ... | | Authors: | Hallberg, B.M, Das, H. | | Deposit date: | 2022-06-28 | | Release date: | 2023-05-10 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Immunoglobulin germline gene polymorphisms influence the function of SARS-CoV-2 neutralizing antibodies.
Immunity, 56, 2023
|
|
8A94
 
 | | SARS CoV2 Spike in the 2-up state in complex with Fab47. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Fab47 (Heavy chain Variable domain), ... | | Authors: | Hallberg, B.M, Das, H. | | Deposit date: | 2022-06-27 | | Release date: | 2023-05-03 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (2.4 Å) | | Cite: | Immunoglobulin germline gene polymorphisms influence the function of SARS-CoV-2 neutralizing antibodies.
Immunity, 56, 2023
|
|
8A95
 
 | | SARS Cov2 Spike RBD in complex with Fab47 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Fab47 (Heavy chain variable domain), ... | | Authors: | Hallberg, B.M, Das, H. | | Deposit date: | 2022-06-27 | | Release date: | 2023-08-16 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.64 Å) | | Cite: | Immunoglobulin germline gene polymorphisms influence the function of SARS-CoV-2 neutralizing antibodies.
Immunity, 56, 2023
|
|
8A96
 
 | | SARS Cov2 Spike RBD in complex with Fab47 | | Descriptor: | Fab47 Heavy chain (variable domain), Fab47 Light chain (variable domain), Spike protein S1 | | Authors: | Hallberg, B.M, Das, H. | | Deposit date: | 2022-06-27 | | Release date: | 2023-05-03 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Immunoglobulin germline gene polymorphisms influence the function of SARS-CoV-2 neutralizing antibodies.
Immunity, 56, 2023
|
|
7B17
 
 | | SARS-CoV-spike RBD bound to two neutralising nanobodies. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, SARS-CoV-2 neutralizing biparatopic nanobody VE,nanobody E from Lama glama,SARS-CoV-2 neutralizing biparatopic nanobody VE,nanobody E from Lama glama, Spike protein S1 | | Authors: | Hallberg, B.M, Das, H. | | Deposit date: | 2020-11-23 | | Release date: | 2021-02-10 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (4.01 Å) | | Cite: | Structure-guided multivalent nanobodies block SARS-CoV-2 infection and suppress mutational escape
Science, 371, 2021
|
|
7B14
 
 | | Nanobody E bound to Spike-RBD in a localized reconstruction | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Nanobody against SARS-CoV-2, Spike protein S1 | | Authors: | Hallberg, B.M, Das, H. | | Deposit date: | 2020-11-23 | | Release date: | 2021-04-28 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.79 Å) | | Cite: | Structure-guided multivalent nanobodies block SARS-CoV-2 infection and suppress mutational escape
Science, 371, 2021
|
|
7B18
 
 | | SARS-CoV-spike bound to two neutralising nanobodies | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Nanobody against SARS-CoV-2 VHH E, ... | | Authors: | Hallberg, B.M, Das, H. | | Deposit date: | 2020-11-24 | | Release date: | 2021-04-28 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (2.62 Å) | | Cite: | Structure-guided multivalent nanobodies block SARS-CoV-2 infection and suppress mutational escape.
Science, 371, 2021
|
|
8V4F
 
 | |
1D7D
 
 | | CYTOCHROME DOMAIN OF CELLOBIOSE DEHYDROGENASE, HP3 FRAGMENT, PH 7.5 | | Descriptor: | 2-(2-{2-[2-(2-METHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHANOL, CADMIUM ION, CELLOBIOSE DEHYDROGENASE, ... | | Authors: | Hallberg, B.M, Bergfors, T, Backbro, K, Divne, C. | | Deposit date: | 1999-10-16 | | Release date: | 2000-10-18 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A new scaffold for binding haem in the cytochrome domain of the extracellular flavocytochrome cellobiose dehydrogenase.
Structure Fold.Des., 8, 2000
|
|
1D7C
 
 | | CYTOCHROME DOMAIN OF CELLOBIOSE DEHYDROGENASE, PH 4.6 | | Descriptor: | 2-(2-{2-[2-(2-METHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHANOL, CADMIUM ION, CELLOBIOSE DEHYDROGENASE, ... | | Authors: | Hallberg, B.M, Bergfors, T, Backbro, K, Divne, C. | | Deposit date: | 1999-10-16 | | Release date: | 2000-10-18 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A new scaffold for binding haem in the cytochrome domain of the extracellular flavocytochrome cellobiose dehydrogenase.
Structure Fold.Des., 8, 2000
|
|
1D7B
 
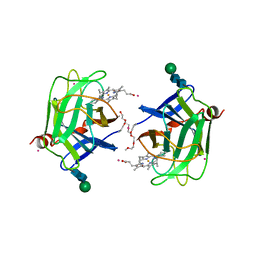 | | CYTOCHROME DOMAIN OF CELLOBIOSE DEHYDROGENASE, PH 7.5 | | Descriptor: | 2-(2-{2-[2-(2-METHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHANOL, CADMIUM ION, CELLOBIOSE DEHYDROGENASE, ... | | Authors: | Hallberg, B.M, Bergfors, T, Backbro, K, Divne, C. | | Deposit date: | 1999-10-16 | | Release date: | 2000-10-18 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A new scaffold for binding haem in the cytochrome domain of the extracellular flavocytochrome cellobiose dehydrogenase.
Structure Fold.Des., 8, 2000
|
|
1KDG
 
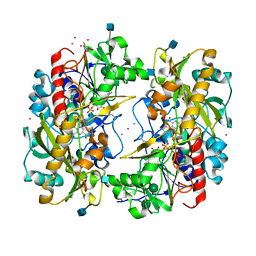 | | Crystal structure of the flavin domain of cellobiose dehydrogenase | | Descriptor: | 2-(ETHYLMERCURI-THIO)-BENZOIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, 6-HYDROXY-FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Hallberg, B.M, Henriksson, G, Pettersson, G, Divne, C. | | Deposit date: | 2001-11-13 | | Release date: | 2002-11-13 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal Structure of the Flavoprotein Domain of the Extracellular Flavocytochrome Cellobiose Dehydrogenase
J.Mol.Biol., 315, 2002
|
|
1TT0
 
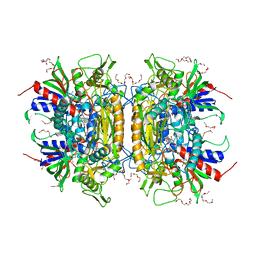 | | Crystal Structure of Pyranose 2-Oxidase | | Descriptor: | ACETATE ION, DODECAETHYLENE GLYCOL, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Hallberg, B.M, Leitner, C, Haltrich, D, Divne, C. | | Deposit date: | 2004-06-21 | | Release date: | 2005-06-21 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of the 270 kDa homotetrameric lignin-degrading enzyme pyranose 2-oxidase
J.Mol.Biol., 341, 2004
|
|
2A98
 
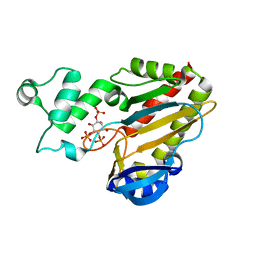 | | Crystal structure of the catalytic domain of human inositol 1,4,5-trisphosphate 3-kinase C | | Descriptor: | D-MYO-INOSITOL-1,4,5-TRIPHOSPHATE, Inositol 1,4,5-trisphosphate 3-kinase C | | Authors: | Hallberg, B.M, Ogg, D, Ehn, M, Graslund, S, Hammarstrom, M, Kotenyova, T, Nilsson-Ehle, P, Nordlund, P, Persson, C, Sagemark, J, Schuler, H, Stenmark, P, Thorsell, A.-G, Arrowsmith, C, Edwards, A, Sundstrom, M, Weigelt, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2005-07-11 | | Release date: | 2005-07-19 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The crystal structure of the catalytic domain of human inositol 1,4,5-trisphosphate 3-kinase C
To be Published
|
|
1NAA
 
 | | Cellobiose Dehydrogenase Flavoprotein Fragment in Complex with Cellobionolactam | | Descriptor: | (2R,3R,4R,5R)-4,5-dihydroxy-2-(hydroxymethyl)-6-oxopiperidin-3-yl beta-D-glucopyranoside, 2-acetamido-2-deoxy-beta-D-glucopyranose, 6-HYDROXY-FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Hallberg, B.M, Henriksson, G, Pettersson, G, Vasella, A, Divne, C. | | Deposit date: | 2002-11-27 | | Release date: | 2003-01-14 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Mechanism of the reductive half-reaction in cellobiose dehydrogenase
J.BIOL.CHEM., 278, 2003
|
|
