1M06
 
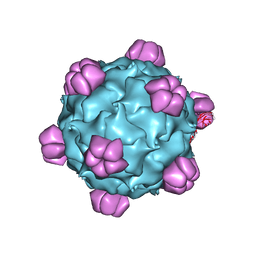 | | Structural Studies of Bacteriophage alpha3 Assembly, X-Ray Crystallography | | Descriptor: | 5'-D(P*(3DR)P*(3DR)P*(3DR)P*(3DR)P*(3DR)P*(3DR)P*(3DR)P*(3DR)P*(3DR)P*(3DR))-3', Capsid Protein, Major spike protein, ... | | Authors: | Bernal, R.A, Hafenstein, S, Olson, N.H, Bowman, V, Chipman, P.R, Baker, T.S, Fane, B.A, Rossmann, M.G. | | Deposit date: | 2002-06-12 | | Release date: | 2002-12-25 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structural Studies of Bacteriophage alpha3 Assembly
J.Mol.Biol., 325, 2003
|
|
1M0F
 
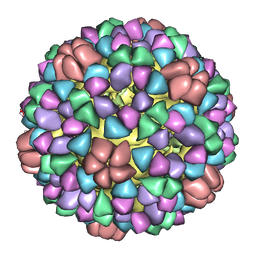 | | Structural Studies of Bacteriophage alpha3 Assembly, Cryo-electron microscopy | | Descriptor: | Capsid Protein F, Major Spike Protein G, Scaffolding Protein B, ... | | Authors: | Bernal, R.A, Hafenstein, S, Olson, N.H, Bowman, V.D, Chipman, P.R, Baker, T.S, Fane, B.A, Rossmann, M.G. | | Deposit date: | 2002-06-12 | | Release date: | 2002-12-25 | | Last modified: | 2024-04-17 | | Method: | ELECTRON MICROSCOPY (16 Å) | | Cite: | Structural Studies of Bacteriophage alpha3 Assembly
J.Mol.Biol., 325, 2003
|
|
3EPC
 
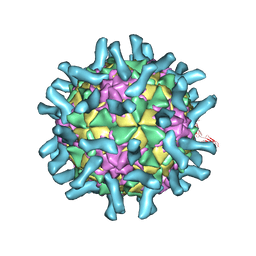 | | CryoEM structure of poliovirus receptor bound to poliovirus type 1 | | Descriptor: | MYRISTIC ACID, Poliovirus receptor, Protein VP1, ... | | Authors: | Zhang, P, Mueller, S, Morais, M.C, Bator, C.M, Bowman, V.D, Hafenstein, S, Wimmer, E, Rossmann, M.G. | | Deposit date: | 2008-09-29 | | Release date: | 2008-11-11 | | Last modified: | 2024-04-17 | | Method: | ELECTRON MICROSCOPY (8 Å) | | Cite: | Crystal structure of CD155 and electron microscopic studies of its complexes with polioviruses.
Proc.Natl.Acad.Sci.USA, 105, 2008
|
|
3EPF
 
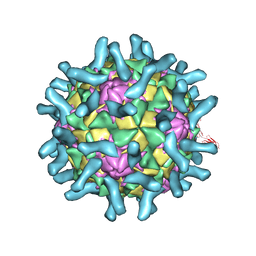 | | CryoEM structure of poliovirus receptor bound to poliovirus type 2 | | Descriptor: | 1[2-CHLORO-4-METHOXY-PHENYL-OXYMETHYL]-4-[2,6-DICHLORO-PHENYL-OXYMETHYL]-BENZENE, MYRISTIC ACID, Poliovirus receptor, ... | | Authors: | Zhang, P, Mueller, S, Morais, M.C, Bator, C.M, Bowman, V.D, Hafenstein, S, Wimmer, E, Rossmann, M.G. | | Deposit date: | 2008-09-29 | | Release date: | 2008-11-11 | | Last modified: | 2024-04-17 | | Method: | ELECTRON MICROSCOPY (9 Å) | | Cite: | Crystal structure of CD155 and electron microscopic studies of its complexes with polioviruses.
Proc.Natl.Acad.Sci.USA, 105, 2008
|
|
3EPD
 
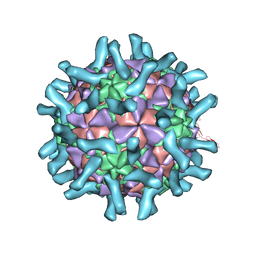 | | CryoEM structure of poliovirus receptor bound to poliovirus type 3 | | Descriptor: | MYRISTIC ACID, Poliovirus Type3 peptide, Poliovirus receptor, ... | | Authors: | Zhang, P, Mueller, S, Morais, M.C, Bator, C.M, Bowman, V.D, Hafenstein, S, Wimmer, E, Rossmann, M.G. | | Deposit date: | 2008-09-29 | | Release date: | 2008-11-11 | | Last modified: | 2024-04-17 | | Method: | ELECTRON MICROSCOPY (9 Å) | | Cite: | Crystal structure of CD155 and electron microscopic studies of its complexes with polioviruses.
Proc.Natl.Acad.Sci.USA, 105, 2008
|
|
4GMP
 
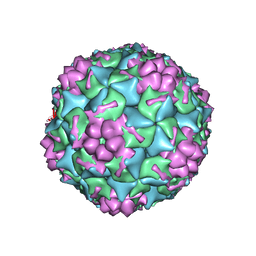 | |
2XGK
 
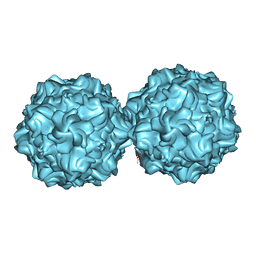 | | Virus like particle of L172W mutant of Minute Virus of Mice - the immunosuppressive strain | | Descriptor: | COAT PROTEIN VP2 | | Authors: | Plevka, P, Hafenstein, S, Tattersall, P, Cotmore, S, Farr, G, D'Abramo, A, Rossmann, M.G. | | Deposit date: | 2010-06-04 | | Release date: | 2011-04-20 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (4.2 Å) | | Cite: | Structure of a Packaging-Defective Mutant of Minute Virus of Mice Indicates that the Genome is Packaged Via a Pore at a 5-Fold Axis.
J.Virol., 85, 2011
|
|
6BSP
 
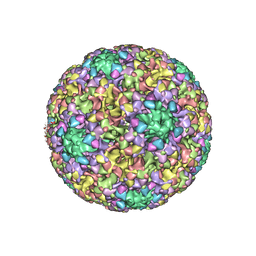 | | High-Resolution Structure Analysis of Antibody V5 and U4 Conformational Epitope on Human Papillomavirus 16 | | Descriptor: | Major capsid protein L1, U4 Heavy chain, U4 Light chain | | Authors: | Guan, J, Bywaters, S.M, Brendle, S.A, Ashley, R.E, Makhov, A.M, Conway, J.F, Christenson, N.D, Hafenstein, S. | | Deposit date: | 2017-12-04 | | Release date: | 2018-02-14 | | Last modified: | 2018-10-24 | | Method: | ELECTRON MICROSCOPY (4.7 Å) | | Cite: | High-Resolution Structure Analysis of Antibody V5 and U4 Conformational Epitopes on Human Papillomavirus 16.
Viruses, 9, 2017
|
|
6DIZ
 
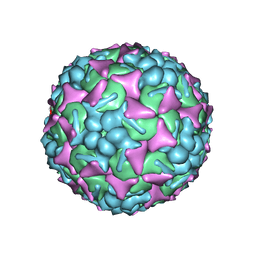 | | EV-A71 strain 11316 complexed with tryptophan dendrimer MADAL_0385 | | Descriptor: | SPHINGOSINE, VP1, VP2, ... | | Authors: | Sun, L, Lee, H, Thibaut, H.J, Rivero-Buceta, E, Martinez-Gualda, B, Delang, L, Leyssen, P, Gago, F, San-Felix, A, Hafenstein, S, Mirabelli, C, Neyts, J. | | Deposit date: | 2018-05-24 | | Release date: | 2019-04-24 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.59 Å) | | Cite: | Viral engagement with host (co-)receptors blocked by a novel class of tryptophan dendrimers that targets the 5-fold-axis of the enterovirus-A71 capsid.
Plos Pathog., 15, 2019
|
|
6BT3
 
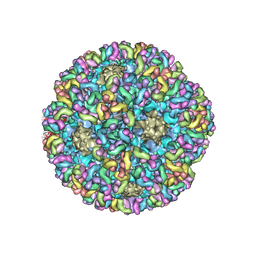 | | High-Resolution Structure Analysis of Antibody V5 Conformational Epitope on Human Papillomavirus 16 | | Descriptor: | Major capsid protein L1, V5 Fab Heavy-chain, V5 Fab Light-chain | | Authors: | Guan, J, Bywaters, S.M, Brendle, S.A, Ashley, R.E, Makhov, A.M, Conway, J.F, Christensen, N.D, Hafenstein, S. | | Deposit date: | 2017-12-05 | | Release date: | 2018-01-17 | | Last modified: | 2018-01-24 | | Method: | ELECTRON MICROSCOPY (4.7 Å) | | Cite: | High-Resolution Structure Analysis of Antibody V5 and U4 Conformational Epitopes on Human Papillomavirus 16.
Viruses, 9, 2017
|
|
4GB3
 
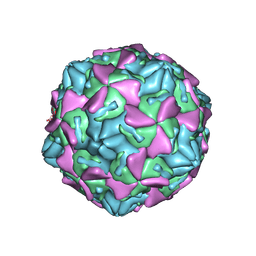 | | Human coxsackievirus B3 strain RD coat protein | | Descriptor: | MYRISTIC ACID, PALMITIC ACID, coat protein 1, ... | | Authors: | Yoder, J.D, Hafenstein, S. | | Deposit date: | 2012-07-26 | | Release date: | 2012-09-26 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.74 Å) | | Cite: | The Crystal Structure of a Coxsackievirus B3-RD Variant and a Refined 9-Angstrom Cryo-Electron Microscopy Reconstruction of the Virus Complexed with Decay-Accelerating Factor (DAF) Provide a New Footprint of DAF on the Virus Surface.
J.Virol., 86, 2012
|
|
2X5I
 
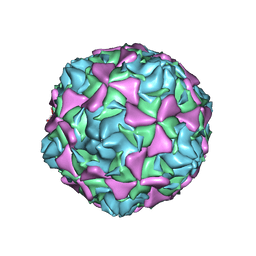 | | Crystal structure echovirus 7 | | Descriptor: | LAURIC ACID, VP1, VP2, ... | | Authors: | Plevka, P, Hafenstein, S, Zhang, Y, Bowman, V.D, Chipman, P.R, Bator, C.M, Rossmann, M.G. | | Deposit date: | 2010-02-08 | | Release date: | 2010-12-22 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Interaction of Decay-Accelerating Factor with Echovirus 7.
J.Virol., 84, 2010
|
|
3J93
 
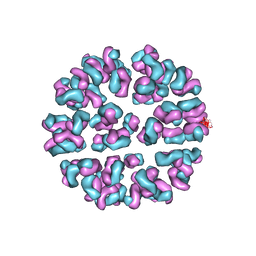 | | Fitting of Fab into the cryoEM density map of EV71 procapsid in complex with Fab22A12 | | Descriptor: | neutralizing antibody 22A12, heavy chain, light chain | | Authors: | Shingler, K.L, Cifuente, J.O, Ashley, R.E, Makhov, A.M, Conway, J.F, Hafenstein, S. | | Deposit date: | 2014-12-02 | | Release date: | 2014-12-24 | | Last modified: | 2018-07-18 | | Method: | ELECTRON MICROSCOPY (8.8 Å) | | Cite: | The enterovirus 71 procapsid binds neutralizing antibodies and rescues virus infection in vitro.
J.Virol., 89, 2015
|
|
3J8Z
 
 | |
3J6L
 
 | | Kinetic and Structural Analysis of Coxsackievirus B3 Receptor Interactions and Formation of the A-particle | | Descriptor: | Coxsackievirus and adenovirus receptor, SULFATE ION | | Authors: | Organtini, L.J, Makhov, A.M, Conway, J.F, Hafenstein, S, Carson, S.D. | | Deposit date: | 2014-03-19 | | Release date: | 2014-04-09 | | Last modified: | 2018-07-18 | | Method: | ELECTRON MICROSCOPY (9 Å) | | Cite: | Kinetic and structural analysis of coxsackievirus b3 receptor interactions and formation of the a-particle.
J.Virol., 88, 2014
|
|
3J6N
 
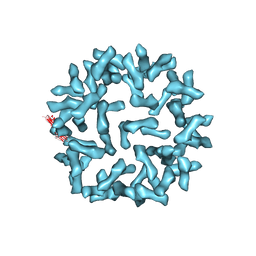 | | Kinetic and Structural Analysis of Coxsackievirus B3 Receptor Interactions and Formation of the A-particle | | Descriptor: | Coxsackie and adenovirus receptor | | Authors: | Organtini, L.J, Makhov, A.M, Conway, J.F, Hafenstein, S, Carson, S.D. | | Deposit date: | 2014-03-19 | | Release date: | 2014-04-09 | | Last modified: | 2018-07-18 | | Method: | ELECTRON MICROSCOPY (9 Å) | | Cite: | Kinetic and structural analysis of coxsackievirus b3 receptor interactions and formation of the a-particle.
J.Virol., 88, 2014
|
|
3J8W
 
 | |
7KZF
 
 | |
7JYI
 
 | | Subparticle Map of ZIKV MR-766 | | Descriptor: | E Glycoprotein, M protein | | Authors: | Jose, J, Hafenstein, S. | | Deposit date: | 2020-08-30 | | Release date: | 2020-09-16 | | Last modified: | 2020-10-14 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Identification of a pocket factor that is critical to Zika virus assembly.
Nat Commun, 11, 2020
|
|
3IYP
 
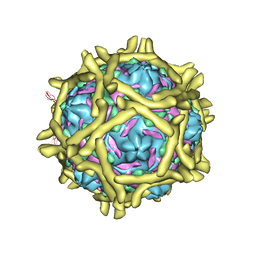 | | The Interaction of Decay-accelerating Factor with Echovirus 7 | | Descriptor: | Capsid protein, Complement decay-accelerating factor, LAURIC ACID, ... | | Authors: | Plevka, P, Hafenstein, S, Zhang, Y, Harris, K.G, Cifuente, J.O, Bowman, V.D, Chipman, P.R, Lin, F, Medof, D.E, Bator, C.M, Rossmann, M.G. | | Deposit date: | 2010-04-07 | | Release date: | 2010-11-24 | | Last modified: | 2024-01-31 | | Method: | ELECTRON MICROSCOPY (7.2 Å) | | Cite: | Interaction of decay-accelerating factor with echovirus 7.
J.Virol., 84, 2010
|
|
3J7E
 
 | | Electron cryo-microscopy of human papillomavirus 16 and H16.V5 Fab fragments | | Descriptor: | H16.V5 Fab heavy chain, H16.V5 Fab light chain | | Authors: | Lee, H, Brendle, S.A, Bywaters, S.M, Christensen, N.D, Hafenstein, S. | | Deposit date: | 2014-06-23 | | Release date: | 2014-11-26 | | Last modified: | 2018-07-18 | | Method: | ELECTRON MICROSCOPY (13.6 Å) | | Cite: | A cryo-electron microscopy study identifies the complete H16.V5 epitope and reveals global conformational changes initiated by binding of the neutralizing antibody fragment.
J.Virol., 89, 2015
|
|
3J3Z
 
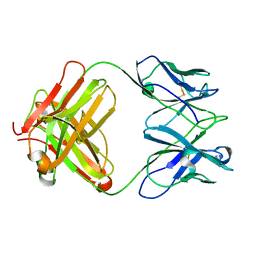 | | Structure of MA28-7 neutralizing antibody Fab fragment from electron cryo-microscopy of enterovirus 71 complexed with a Fab fragment | | Descriptor: | MA28-7 neutralizing antibody heavy chain, MA28-7 neutralizing antibody light chain | | Authors: | Lee, H, Cifuente, J.O, Ashley, R.E, Conway, J.F, Makhov, A.M, Tano, Y, Shimizu, H, Nishimura, Y, Hafenstein, S. | | Deposit date: | 2013-05-21 | | Release date: | 2013-08-28 | | Last modified: | 2018-07-18 | | Method: | ELECTRON MICROSCOPY (23.4 Å) | | Cite: | A strain-specific epitope of enterovirus 71 identified by cryo-electron microscopy of the complex with fab from neutralizing antibody.
J.Virol., 87, 2013
|
|
3J6M
 
 | | Kinetic and Structural Analysis of Coxsackievirus B3 Receptor Interactions and Formation of the A-particle | | Descriptor: | Coxsackievirus and adenovirus receptor | | Authors: | Organtini, L.J, Makhov, A.M, Conway, J.F, Hafenstein, S, Carson, S.D. | | Deposit date: | 2014-03-19 | | Release date: | 2014-04-09 | | Last modified: | 2018-07-18 | | Method: | ELECTRON MICROSCOPY (9 Å) | | Cite: | Kinetic and structural analysis of coxsackievirus b3 receptor interactions and formation of the a-particle.
J.Virol., 88, 2014
|
|
3J6O
 
 | | Kinetic and Structural Analysis of Coxsackievirus B3 Receptor Interactions and Formation of the A-particle | | Descriptor: | Coxsackie and adenovirus receptor | | Authors: | Organtini, L.J, Makhov, A.M, Conway, J.F, Hafenstein, S, Carson, S.D. | | Deposit date: | 2014-03-19 | | Release date: | 2014-04-09 | | Last modified: | 2018-07-18 | | Method: | ELECTRON MICROSCOPY (9 Å) | | Cite: | Kinetic and structural analysis of coxsackievirus b3 receptor interactions and formation of the a-particle.
J.Virol., 88, 2014
|
|
3J8V
 
 | |
