2ORM
 
 | | Crystal Structure of the 4-Oxalocrotonate Tautomerase Homologue DmpI from Helicobacter pylori. | | Descriptor: | Probable tautomerase HP0924 | | Authors: | Hackert, M.L, Whitman, C.P, Almrud, J.J, Dasgupta, R, Czerwinski, R.M, Kern, A.D. | | Deposit date: | 2007-02-03 | | Release date: | 2008-02-12 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Kinetic and structural characterization of DmpI from Helicobacter pylori and Archaeoglobus fulgidus, two 4-oxalocrotonate tautomerase family members.
Bioorg.Chem., 38, 2010
|
|
1HLB
 
 | |
3M20
 
 | | Crystal structure of DmpI from Archaeoglobus fulgidus determined to 2.37 Angstroms resolution | | Descriptor: | 4-oxalocrotonate tautomerase, putative | | Authors: | Hackert, M.L, Whitman, C.P, Almrud, J.J, Dasgupta, R, Kern, A.D. | | Deposit date: | 2010-03-06 | | Release date: | 2010-09-01 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Kinetic and structural characterization of DmpI from Helicobacter pylori and Archaeoglobus fulgidus, two 4-oxalocrotonate tautomerase family members.
Bioorg.Chem., 38, 2010
|
|
3M21
 
 | | Crystal structure of DmpI from Helicobacter pylori Determined to 1.9 Angstroms resolution | | Descriptor: | Probable tautomerase HP_0924 | | Authors: | Hackert, M.L, Whitman, C.P, Almrud, J.J, Dasgupta, R, Kern, A.D, Czerwinski, R.M. | | Deposit date: | 2010-03-06 | | Release date: | 2010-09-01 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Kinetic and structural characterization of DmpI from Helicobacter pylori and Archaeoglobus fulgidus, two 4-oxalocrotonate tautomerase family members.
Bioorg.Chem., 38, 2010
|
|
1ITH
 
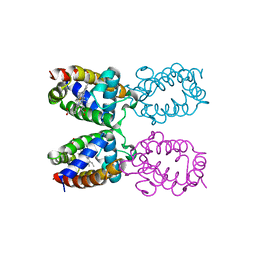 | | STRUCTURE DETERMINATION AND REFINEMENT OF HOMOTETRAMERIC HEMOGLOBIN FROM URECHIS CAUPO AT 2.5 ANGSTROMS RESOLUTION | | Descriptor: | CYANIDE ION, HEMOGLOBIN (CYANO MET), PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Hackert, M, Kolatkar, P, Ernst, S.R, Ogata, C.M, Hendrickson, W.A, Merritt, E.A, Phizackerley, R.P. | | Deposit date: | 1991-12-03 | | Release date: | 1993-10-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure determination and refinement of homotetrameric hemoglobin from Urechis caupo at 2.5 A resolution.
Acta Crystallogr.,Sect.B, 48, 1992
|
|
1ORD
 
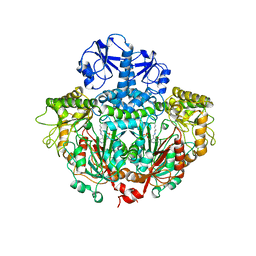 | | CRYSTALLOGRAPHIC STRUCTURE OF A PLP-DEPENDENT ORNITHINE DECARBOXYLASE FROM LACTOBACILLUS 30A TO 3.1 ANGSTROMS RESOLUTION | | Descriptor: | ORNITHINE DECARBOXYLASE, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Hackert, M.L, Momany, C, Ernst, S, Ghosh, R. | | Deposit date: | 1995-02-08 | | Release date: | 1995-09-15 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystallographic structure of a PLP-dependent ornithine decarboxylase from Lactobacillus 30a to 3.0 A resolution.
J.Mol.Biol., 252, 1995
|
|
2OP8
 
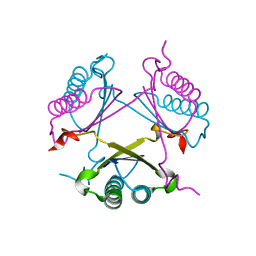 | |
1HLM
 
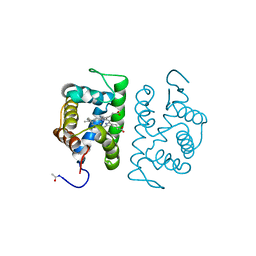 | |
2OPA
 
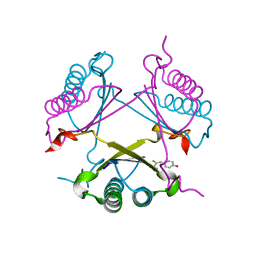 | | YwhB binary complex with 2-Fluoro-p-hydroxycinnamate | | Descriptor: | 2-FLUORO-3-(4-HYDROXYPHENYL)-2E-PROPENEOATE, Probable tautomerase ywhB | | Authors: | Hackert, M.L, Whitman, C.P, Almrud, J.J. | | Deposit date: | 2007-01-28 | | Release date: | 2008-02-05 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The Crystal Structure of YwhB, a 4-Oxalocrotonate Tautomerase Homologue from Bacillus subtilis: the Structural Basis for Catalysis, Inhibition, and Reaction Stereoselectivity.
TO BE PUBLISHED
|
|
5MU8
 
 | | HUMAN TNF-ALPHA IN COMPLEX WITH JNJ525 | | Descriptor: | Tumor necrosis factor, ~{N}4-(phenylmethyl)-~{N}4-[2-[3-(2-piperazin-1-ylpyrimidin-5-yl)phenyl]phenyl]pyrimidine-2,4-diamine | | Authors: | Blevitt, J.M, Hack, M.D, Herman, K.L, Jackson, P.F, Krawczuk, P.J, Lebsack, A.D, Liu, A.X, Mirzadegan, T, Nelen, M.I, Patrick, A.P, Steinbacher, S, Milla, M.E, Lumb, K.J. | | Deposit date: | 2017-01-12 | | Release date: | 2017-03-29 | | Last modified: | 2019-02-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural Basis of Small-Molecule Aggregate Induced Inhibition of a Protein-Protein Interaction.
J. Med. Chem., 60, 2017
|
|
6WPP
 
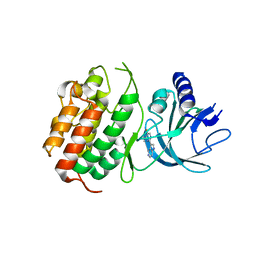 | | HUMAN NIK IN COMPLEX WITH LIGAND COMPOUND X | | Descriptor: | 5-fluoro-N-methyl-2-(7H-pyrrolo[2,3-d]pyrimidin-5-yl)pyrimidin-4-amine, Mitogen-activated protein kinase kinase kinase 14 | | Authors: | Shih, A.Y, Hack, M, Mirzadegan, T. | | Deposit date: | 2020-04-27 | | Release date: | 2020-08-26 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Impact of Protein Preparation on Resulting Accuracy of FEP Calculations.
J.Chem.Inf.Model., 60, 2020
|
|
1TM2
 
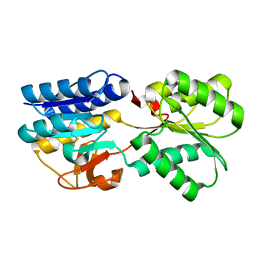 | | Crystal Structure of the apo form of the Salmonella typhimurium AI-2 receptor LsrB | | Descriptor: | sugar transport protein | | Authors: | Miller, S.T, Xavier, K.B, Campagna, S.R, Taga, M.E, Semmelhack, M.F, Bassler, B.L, Hughson, F.M. | | Deposit date: | 2004-06-10 | | Release date: | 2004-09-28 | | Last modified: | 2018-01-31 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Salmonella typhimurium Recognizes a Chemically Distinct Form of the Bacterial Quorum-Sensing Signal AI-2
Mol.Cell, 15, 2004
|
|
7PDZ
 
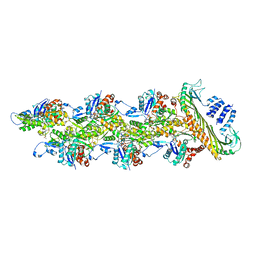 | | Structure of capping protein bound to the barbed end of a cytoplasmic actin filament | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, cytoplasmic 1, ... | | Authors: | Funk, J, Merino, F, Schacks, M, Rottner, K, Raunser, S, Bieling, P. | | Deposit date: | 2021-08-09 | | Release date: | 2021-09-01 | | Last modified: | 2021-10-06 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | A barbed end interference mechanism reveals how capping protein promotes nucleation in branched actin networks.
Nat Commun, 12, 2021
|
|
7ODC
 
 | | CRYSTAL STRUCTURE ORNITHINE DECARBOXYLASE FROM MOUSE, TRUNCATED 37 RESIDUES FROM THE C-TERMINUS, TO 1.6 ANGSTROM RESOLUTION | | Descriptor: | PROTEIN (ORNITHINE DECARBOXYLASE), PYRIDOXAL-5'-PHOSPHATE | | Authors: | Kern, A.D, Oliveira, M.A, Coffino, P, Hackert, M.L. | | Deposit date: | 1999-03-03 | | Release date: | 1999-10-22 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure of mammalian ornithine decarboxylase at 1.6 A resolution: stereochemical implications of PLP-dependent amino acid decarboxylases.
Structure Fold.Des., 7, 1999
|
|
3LDH
 
 | | A comparison of the structures of apo dogfish m4 lactate dehydrogenase and its ternary complexes | | Descriptor: | LACTATE DEHYDROGENASE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, PYRUVIC ACID | | Authors: | White, J.L, Hackert, M.L, Buehner, M, Adams, M.J, Ford, G.C, Lentzjunior, P.J, Smiley, I.E, Steindel, S.J, Rossmann, M.G. | | Deposit date: | 1974-06-06 | | Release date: | 1977-04-29 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | A comparison of the structures of apo dogfish M4 lactate dehydrogenase and its ternary complexes.
J.Mol.Biol., 102, 1976
|
|
1TJY
 
 | | Crystal Structure of Salmonella typhimurium AI-2 receptor LsrB in complex with R-THMF | | Descriptor: | (2R,4S)-2-methyl-2,3,3,4-tetrahydroxytetrahydrofuran, sugar transport protein | | Authors: | Miller, S.T, Xavier, K.B, Campagna, S.R, Taga, M.E, Semmelhack, M.F, Bassler, B.L, Hughson, F.M. | | Deposit date: | 2004-06-07 | | Release date: | 2004-09-28 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Salmonella typhimurium Recognizes a Chemically Distinct Form of the Bacterial Quorum-Sensing Signal AI-2
Mol.Cell, 15, 2004
|
|
4LHP
 
 | | Crystal Structure of Native FG41Malonate Semialdehyde Decarboxylase | | Descriptor: | FG41 Malonate Semialdehyde Decarboxylase, PHOSPHATE ION, SULFATE ION | | Authors: | Guo, Y, Serrano, H, Poelarends, G.J, Johnson Jr, W.H, Hackert, M.L, Whitman, C.P. | | Deposit date: | 2013-07-01 | | Release date: | 2013-07-24 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Kinetic, Mutational, and Structural Analysis of Malonate Semialdehyde Decarboxylase from Coryneform Bacterium Strain FG41: Mechanistic Implications for the Decarboxylase and Hydratase Activities.
Biochemistry, 52, 2013
|
|
4LHO
 
 | | Crystal Structure of FG41Malonate Semialdehyde Decarboxylase inhibited by 3-bromopropiolate | | Descriptor: | 3-chloro-3-oxopropanoic acid, FG41 Malonate Semialdehyde Decarboxylase, PHOSPHATE ION | | Authors: | Guo, Y, Serrano, H, Poelarends, G.J, Johnson Jr, W.H, Hackert, M.L, Whitman, C.P. | | Deposit date: | 2013-07-01 | | Release date: | 2013-07-24 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.224 Å) | | Cite: | Kinetic, Mutational, and Structural Analysis of Malonate Semialdehyde Decarboxylase from Coryneform Bacterium Strain FG41: Mechanistic Implications for the Decarboxylase and Hydratase Activities.
Biochemistry, 52, 2013
|
|
6V0T
 
 | | Crystal Structure of Catalytic Subunit of Bovine Pyruvate Dehydrogenase Phosphatase 1 - Catalytic Domain | | Descriptor: | MANGANESE (II) ION, SULFATE ION, [Pyruvate dehydrogenase [acetyl-transferring]]-phosphatase 1, ... | | Authors: | Guo, Y, Qiu, W, Ernst, S.R, Carroll, D.W, Hackert, M.L. | | Deposit date: | 2019-11-19 | | Release date: | 2019-12-18 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of the catalytic subunit of bovine pyruvate dehydrogenase phosphatase.
Acta Crystallogr.,Sect.F, 76, 2020
|
|
4OTA
 
 | |
1MFI
 
 | | CRYSTAL STRUCTURE OF MACROPHAGE MIGRATION INHIBITORY FACTOR COMPLEXED WITH (E)-2-FLUORO-P-HYDROXYCINNAMATE | | Descriptor: | 2-FLUORO-3-(4-HYDROXYPHENYL)-2E-PROPENEOATE, PROTEIN (MACROPHAGE MIGRATION INHIBITORY FACTOR) | | Authors: | Taylor, A.B, Johnson Jr, W.H, Czerwinski, R.M, Whitman, C.P, Hackert, M.L. | | Deposit date: | 1998-08-12 | | Release date: | 1999-06-22 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of macrophage migration inhibitory factor complexed with (E)-2-fluoro-p-hydroxycinnamate at 1.8 A resolution: implications for enzymatic catalysis and inhibition.
Biochemistry, 38, 1999
|
|
2GDG
 
 | |
1PYA
 
 | |
2FM7
 
 | |
1E2O
 
 | | CATALYTIC DOMAIN FROM DIHYDROLIPOAMIDE SUCCINYLTRANSFERASE | | Descriptor: | DIHYDROLIPOAMIDE SUCCINYLTRANSFERASE, SULFATE ION | | Authors: | Knapp, J.E, Mitchell, D.T, Yazdi, M.A, Ernst, S.R, Reed, L.J, Hackert, M.L. | | Deposit date: | 1998-05-26 | | Release date: | 1998-12-02 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of the truncated cubic core component of the Escherichia coli 2-oxoglutarate dehydrogenase multienzyme complex.
J.Mol.Biol., 280, 1998
|
|
