1N6R
 
 | | Crystal Structure of Human Rab5a A30L mutant complex with GppNHp | | 分子名称: | BETA-MERCAPTOETHANOL, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, ... | | 著者 | Zhu, G, Liu, J, Terzyan, S, Zhai, P, Li, G, Zhang, X.C. | | 登録日 | 2002-11-11 | | 公開日 | 2002-11-27 | | 最終更新日 | 2021-10-27 | | 実験手法 | X-RAY DIFFRACTION (1.55 Å) | | 主引用文献 | High Resolution Crystal Structures of Human Rab5a and Five Mutants with Substitutions in the Catalytically Important Phosphate-Binding Loop
J.Biol.Chem., 278, 2003
|
|
7XC8
 
 | | Crystal structure of cotton alpha-like expansin GhEXLA1 | | 分子名称: | Beta-expansin, DI(HYDROXYETHYL)ETHER, GLYCEROL | | 著者 | Zhao, F, Men, S, Xue, Y, Tu, L.L, Yin, P, Zhang, X.L. | | 登録日 | 2022-03-23 | | 公開日 | 2023-05-17 | | 実験手法 | X-RAY DIFFRACTION (2.55 Å) | | 主引用文献 | Crystal structure of cotton alpha-like expansin GhEXLA1
To Be Published
|
|
6LJU
 
 | | Crystal structure of human FABP4 in complex with a novel inhibitor | | 分子名称: | 1,2-ETHANEDIOL, 2-[[3-chloranyl-4-(methylamino)-2-phenyl-phenyl]amino]benzoic acid, Fatty acid-binding protein, ... | | 著者 | Su, H.X, Zhang, X.L, Li, M.J, Xu, Y.C. | | 登録日 | 2019-12-17 | | 公開日 | 2020-04-15 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Exploration of Fragment Binding Poses Leading to Efficient Discovery of Highly Potent and Orally Effective Inhibitors of FABP4 for Anti-inflammation.
J.Med.Chem., 63, 2020
|
|
6LJX
 
 | | Crystal structure of human FABP4 in complex with a novel inhibitor | | 分子名称: | 2-phenylazanylbenzoic acid, Fatty acid-binding protein, adipocyte | | 著者 | Su, H.X, Zhang, X.L, Li, M.J, Xu, Y.C. | | 登録日 | 2019-12-17 | | 公開日 | 2020-04-15 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | Exploration of Fragment Binding Poses Leading to Efficient Discovery of Highly Potent and Orally Effective Inhibitors of FABP4 for Anti-inflammation.
J.Med.Chem., 63, 2020
|
|
7XU2
 
 | | Structure of SARS-CoV-2 Spike Protein with Engineered x3 Disulfide (x3(D427C, V987C) and single Arg S1/S2 cleavage site), Locked-2 Conformation | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BILIVERDINE IX ALPHA, ... | | 著者 | Qu, K, Chen, Q, Ciazynska, K.A, Liu, B, Zhang, X, Wang, J, He, Y, Guan, J, He, J, Liu, T, Zhang, X, Carter, A.P, Xiong, X, Briggs, J.A.G. | | 登録日 | 2022-05-18 | | 公開日 | 2022-07-20 | | 最終更新日 | 2022-08-17 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | Engineered disulfide reveals structural dynamics of locked SARS-CoV-2 spike.
Plos Pathog., 18, 2022
|
|
7XU0
 
 | | Structure of SARS-CoV-2 Spike Protein with Engineered x3 Disulfide (x3(D427C, V987C) and single Arg S1/S2 cleavage site), Locked-211 Conformation | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BILIVERDINE IX ALPHA, ... | | 著者 | Qu, K, Chen, Q, Ciazynska, K.A, Liu, B, Zhang, X, Wang, J, He, Y, Guan, J, He, J, Liu, T, Zhang, X, Carter, A.P, Xiong, X, Briggs, J.A.G. | | 登録日 | 2022-05-18 | | 公開日 | 2022-07-20 | | 最終更新日 | 2022-08-17 | | 実験手法 | ELECTRON MICROSCOPY (2.9 Å) | | 主引用文献 | Engineered disulfide reveals structural dynamics of locked SARS-CoV-2 spike.
Plos Pathog., 18, 2022
|
|
6LJV
 
 | | Crystal structure of human FABP4 in complex with a novel inhibitor | | 分子名称: | 2-[[3-chloranyl-2-(2,3-dihydro-1-benzofuran-5-yl)phenyl]amino]benzoic acid, Fatty acid-binding protein, adipocyte | | 著者 | Su, H.X, Zhang, X.L, Li, M.J, Xu, Y.C. | | 登録日 | 2019-12-17 | | 公開日 | 2020-04-15 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.401 Å) | | 主引用文献 | Exploration of Fragment Binding Poses Leading to Efficient Discovery of Highly Potent and Orally Effective Inhibitors of FABP4 for Anti-inflammation.
J.Med.Chem., 63, 2020
|
|
6LJW
 
 | | Crystal structure of human FABP4 in complex with a novel inhibitor | | 分子名称: | 1,2-ETHANEDIOL, 2-phenylazanylbenzoic acid, Fatty acid-binding protein, ... | | 著者 | Su, H.X, Zhang, X.L, Li, M.J, Xu, Y.C. | | 登録日 | 2019-12-17 | | 公開日 | 2020-04-15 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | Exploration of Fragment Binding Poses Leading to Efficient Discovery of Highly Potent and Orally Effective Inhibitors of FABP4 for Anti-inflammation.
J.Med.Chem., 63, 2020
|
|
7Y1A
 
 | | Lateral hexamer | | 分子名称: | B-phycoerythrin beta chain, LRH, PHYCOERYTHROBILIN, ... | | 著者 | You, X, Zhang, X, Cheng, J, Xiao, Y.N, Sun, S, Sui, S.F. | | 登録日 | 2022-06-07 | | 公開日 | 2023-01-18 | | 最終更新日 | 2023-04-19 | | 実験手法 | ELECTRON MICROSCOPY (6.3 Å) | | 主引用文献 | In situ structure of the red algal phycobilisome-PSII-PSI-LHC megacomplex.
Nature, 616, 2023
|
|
7Y4L
 
 | | PBS of PBS-PSII-PSI-LHCs from Porphyridium purpureum. | | 分子名称: | Allophycocyanin alpha subunit, Allophycocyanin beta 18 subunit, Allophycocyanin beta subunit, ... | | 著者 | You, X, Zhang, X, Cheng, J, Xiao, Y.N, Sun, S, Sui, S.F. | | 登録日 | 2022-06-15 | | 公開日 | 2023-01-18 | | 最終更新日 | 2023-04-19 | | 実験手法 | ELECTRON MICROSCOPY (3.3 Å) | | 主引用文献 | In situ structure of the red algal phycobilisome-PSII-PSI-LHC megacomplex.
Nature, 616, 2023
|
|
3OZZ
 
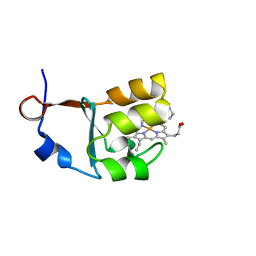 | |
4AIY
 
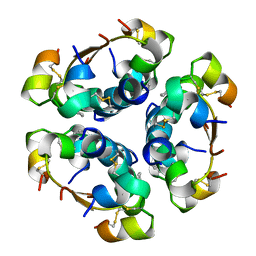 | | R6 HUMAN INSULIN HEXAMER (SYMMETRIC), NMR, 'GREEN' SUBSTATE, AVERAGE STRUCTURE | | 分子名称: | PHENOL, PROTEIN (INSULIN) | | 著者 | O'Donoghue, S.I, Chang, X, Abseher, R, Nilges, M, Led, J.J. | | 登録日 | 1998-12-29 | | 公開日 | 2000-02-28 | | 最終更新日 | 2023-12-27 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Unraveling the symmetry ambiguity in a hexamer: calculation of the R6 human insulin structure.
J.Biomol.NMR, 16, 2000
|
|
7WQV
 
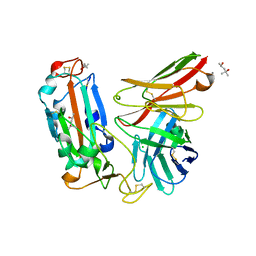 | | Crystal structure of a neutralizing monoclonal antibody (Ab08) in complex with SARS-CoV-2 receptor-binding domain (RBD) | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, Ab08, ... | | 著者 | Zha, J, Meng, L, Zhang, X, Li, D. | | 登録日 | 2022-01-26 | | 公開日 | 2023-01-25 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | A Spike-destructing human antibody effectively neutralizes Omicron-included SARS-CoV-2 variants with therapeutic efficacy.
Plos Pathog., 19, 2023
|
|
2J48
 
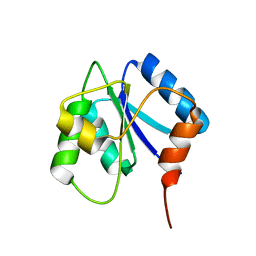 | |
7XRG
 
 | |
6IYA
 
 | |
5ZC3
 
 | |
7VVS
 
 | |
6K8P
 
 | |
2GZY
 
 | |
7Y7A
 
 | | In situ double-PBS-PSII-PSI-LHCs megacomplex from Porphyridium purpureum. | | 分子名称: | (1R,2S)-4-{(1E,3E,5E,7E,9E,11E,13E,15E,17E)-18-[(4S)-4-hydroxy-2,6,6-trimethylcyclohex-1-en-1-yl]-3,7,12,16-tetramethyloctadeca-1,3,5,7,9,11,13,15,17-nonaen-1-yl}-2,5,5-trimethylcyclohex-3-en-1-ol, (2S)-2,3-dihydroxypropyl octadecanoate, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, ... | | 著者 | You, X, Zhang, X, Cheng, J, Xiao, Y.N, Sun, S, Sui, S.F. | | 登録日 | 2022-06-22 | | 公開日 | 2023-02-08 | | 最終更新日 | 2024-07-03 | | 実験手法 | ELECTRON MICROSCOPY (4.3 Å) | | 主引用文献 | In situ structure of the red algal phycobilisome-PSII-PSI-LHC megacomplex.
Nature, 616, 2023
|
|
7Y5E
 
 | | In situ single-PBS-PSII-PSI-LHCs megacomplex. | | 分子名称: | (1R,2S)-4-{(1E,3E,5E,7E,9E,11E,13E,15E,17E)-18-[(4S)-4-hydroxy-2,6,6-trimethylcyclohex-1-en-1-yl]-3,7,12,16-tetramethyloctadeca-1,3,5,7,9,11,13,15,17-nonaen-1-yl}-2,5,5-trimethylcyclohex-3-en-1-ol, (2S)-2,3-dihydroxypropyl octadecanoate, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, ... | | 著者 | You, X, Zhang, X, Cheng, J, Xiao, Y.N, Sui, S.F. | | 登録日 | 2022-06-17 | | 公開日 | 2023-02-01 | | 最終更新日 | 2024-07-03 | | 実験手法 | ELECTRON MICROSCOPY (3.3 Å) | | 主引用文献 | In situ structure of the red algal phycobilisome-PSII-PSI-LHC megacomplex.
Nature, 616, 2023
|
|
6LZ3
 
 | |
8WRB
 
 | | Lysophosphatidylserine receptor GPR34-Gi complex | | 分子名称: | Antibody fragment scFv16, CHOLESTEROL, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | 著者 | Gong, W, Liu, G, Li, X, Wang, Y, Zhang, X. | | 登録日 | 2023-10-13 | | 公開日 | 2023-11-08 | | 最終更新日 | 2023-12-20 | | 実験手法 | ELECTRON MICROSCOPY (2.91 Å) | | 主引用文献 | Structural basis for ligand recognition and signaling of the lysophosphatidylserine receptors GPR34 and GPR174.
Plos Biol., 21, 2023
|
|
8W88
 
 | | Cryo-EM structure of the SEP363856-bound TAAR1-Gs complex | | 分子名称: | 1-[(7~{S})-5,7-dihydro-4~{H}-thieno[2,3-c]pyran-7-yl]-~{N}-methyl-methanamine, CHOLESTEROL, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | 著者 | Liu, H, Zheng, Y, Wang, Y, Wang, Y, He, X, Xu, P, Huang, S, Yuan, Q, Zhang, X, Wang, S, Xu, H.E, Xu, F. | | 登録日 | 2023-09-01 | | 公開日 | 2023-11-22 | | 最終更新日 | 2024-01-03 | | 実験手法 | ELECTRON MICROSCOPY (2.6 Å) | | 主引用文献 | Recognition of methamphetamine and other amines by trace amine receptor TAAR1.
Nature, 624, 2023
|
|
