4O9F
 
 | |
6IMD
 
 | | Crystal structure of PDE4D complexed with a novel inhibitor | | Descriptor: | 1,2-ETHANEDIOL, 6,7-dimethoxy-3,4-dihydroisoquinoline-2(1H)-carbaldehyde, MAGNESIUM ION, ... | | Authors: | Zhang, X, Su, H, Xu, Y. | | Deposit date: | 2018-10-22 | | Release date: | 2019-10-23 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.499 Å) | | Cite: | Structure-Aided Identification and Optimization of Tetrahydro-isoquinolines as Novel PDE4 Inhibitors Leading to Discovery of an Effective Antipsoriasis Agent.
J.Med.Chem., 62, 2019
|
|
4Q2M
 
 | |
4Q2L
 
 | |
5YB3
 
 | | Crystal structure of HP23L/N36 | | Descriptor: | Envelope glycoprotein, HP23L | | Authors: | Zhang, X, Wang, X, He, Y. | | Deposit date: | 2017-09-03 | | Release date: | 2018-02-28 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.043 Å) | | Cite: | Structural Insights into the Mechanisms of Action of Short-Peptide HIV-1 Fusion Inhibitors Targeting the Gp41 Pocket
Front Cell Infect Microbiol, 8, 2018
|
|
4GJF
 
 | |
4GJG
 
 | |
4PR3
 
 | | Crystal structure of Brucella melitensis 5'-methylthioadenosine/S-adenosylhomocysteine nucleosidase | | Descriptor: | 5'-methylthioadenosine nucleosidase / s-adenosylhomocysteine nucleosidase, ADENINE, GLYCEROL, ... | | Authors: | Zhang, X.C, Kang, X.S, Zhao, Y, Jiang, D.H, Li, X.M, Chen, Z.L. | | Deposit date: | 2014-03-05 | | Release date: | 2014-04-30 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.606 Å) | | Cite: | Crystal structure and biochemical studies of Brucella melitensis 5'-methylthioadenosine/S-adenosylhomocysteine nucleosidase
Biochem.Biophys.Res.Commun., 446, 2014
|
|
3RYT
 
 | | The Plexin A1 intracellular region in complex with Rac1 | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, Plexin-A1, ... | | Authors: | Zhang, X, He, H. | | Deposit date: | 2011-05-11 | | Release date: | 2012-01-18 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.582 Å) | | Cite: | Plexins Are GTPase-Activating Proteins for Rap and Are Activated by Induced Dimerization.
Sci.Signal., 5, 2012
|
|
1E32
 
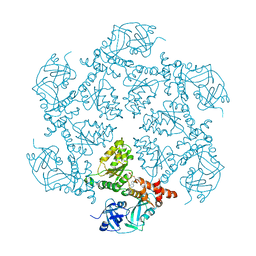 | | Structure of the N-Terminal domain and the D1 AAA domain of membrane fusion ATPase p97 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, P97 | | Authors: | Zhang, X, Shaw, A, Bates, P.A, Gorman, M.A, Kondo, H, Dokurno, P, Leonard M, G, Sternberg, J.E, Freemont, P.S. | | Deposit date: | 2000-06-05 | | Release date: | 2001-05-31 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure of the Aaa ATPase P97
Mol.Cell, 6, 2000
|
|
1UON
 
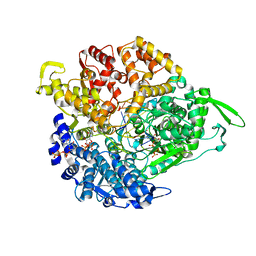 | | REOVIRUS POLYMERASE LAMBDA-3 LOCALIZED BY ELECTRON CRYOMICROSCOPY OF VIRIONS AT 7.6-A RESOLUTION | | Descriptor: | 3'-DEOXY-CYTIDINE-5'-TRIPHOSPHATE, 5'-R(*GP*GP*GP*GP*GP*)-3', 5'-R(*UP*AP*GP*CP*CP*CP*CP*CP*)-3', ... | | Authors: | Zhang, X, Walker, S.B, Chipman, P.R, Nibert, M.L, Baker, T.S. | | Deposit date: | 2003-09-21 | | Release date: | 2003-11-13 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (7.6 Å) | | Cite: | Reovirus Polymerase Lambda3 Localized by Cryo-Electron Microscopy of Virions at a Resolution of 7.6 A
Nat.Struct.Biol., 10, 2003
|
|
6IMM
 
 | | Cryo-EM structure of an alphavirus, Sindbis virus | | Descriptor: | Assembly protein E3, Octadecane, Spike glycoprotein E1, ... | | Authors: | Zhang, X, Ma, J, Chen, L. | | Deposit date: | 2018-10-23 | | Release date: | 2019-03-13 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Implication for alphavirus host-cell entry and assembly indicated by a 3.5 angstrom resolution cryo-EM structure.
Nat Commun, 9, 2018
|
|
6IM6
 
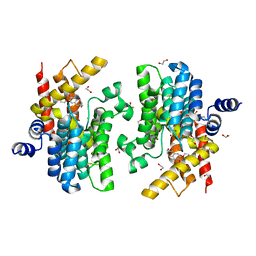 | | Crystal structure of PDE4D complexed with a novel inhibitor | | Descriptor: | 1,2-ETHANEDIOL, 7-ethoxy-6-methoxy-3,4-dihydroisoquinoline-2(1H)-carbaldehyde, MAGNESIUM ION, ... | | Authors: | Zhang, X.L, Su, H.X, Xu, Y.C. | | Deposit date: | 2018-10-22 | | Release date: | 2019-10-23 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.702 Å) | | Cite: | Structure-Aided Identification and Optimization of Tetrahydro-isoquinolines as Novel PDE4 Inhibitors Leading to Discovery of an Effective Antipsoriasis Agent.
J.Med.Chem., 62, 2019
|
|
5YC0
 
 | | Crystal structure of LP-46/N44 | | Descriptor: | Envelope glycoprotein, LP-46 | | Authors: | Zhang, X, Wang, X, He, Y. | | Deposit date: | 2017-09-05 | | Release date: | 2018-02-14 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Exceptional potency and structural basis of a T1249-derived lipopeptide fusion inhibitor against HIV-1, HIV-2, and simian immunodeficiency virus
J. Biol. Chem., 293, 2018
|
|
7YSK
 
 | | Crystal structure of D-Cysteine desulfhydrase from Pectobacterium atrosepticum | | Descriptor: | D-Cysteine desulfhydrase | | Authors: | Zhang, X, Wang, L, Xu, X, Xing, X, Zhou, J. | | Deposit date: | 2022-08-12 | | Release date: | 2022-12-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Characterization and structural basis of D-cysteine desulfhydrase from Pectobacterium atrosepticum
Tetrahedron, 2022
|
|
7YSL
 
 | | Crystal structure of D-Cysteine desulfhydrase with a trapped PLP-pyruvate geminal diamine | | Descriptor: | 1,2-ETHANEDIOL, D-Cysteine desulfhydrase, FORMIC ACID | | Authors: | Zhang, X, Wang, L, Xu, X, Xing, X, Zhou, J. | | Deposit date: | 2022-08-12 | | Release date: | 2022-12-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Characterization and structural basis of D-cysteine desulfhydrase from Pectobacterium atrosepticum
Tetrahedron, 2022
|
|
7DYA
 
 | | Crystal structure of TmFtn with calcium ions | | Descriptor: | CALCIUM ION, FE (III) ION, Ferritin | | Authors: | Zhang, X, Zhao, G. | | Deposit date: | 2021-01-20 | | Release date: | 2021-09-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.197 Å) | | Cite: | Protein interface redesign facilitates the transformation of nanocage building blocks to 1D and 2D nanomaterials.
Nat Commun, 12, 2021
|
|
8H13
 
 | | Structure of SARS-CoV-1 Spike Protein with Engineered x2 Disulfide (G400C and V969C), Closed Conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Zhang, X, Li, Z, Liu, Y, Wang, J, Fu, L, Wang, P, He, J, Xiong, X. | | Deposit date: | 2022-09-30 | | Release date: | 2022-10-19 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (4.05 Å) | | Cite: | Disulfide stabilization reveals conserved dynamic features between SARS-CoV-1 and SARS-CoV-2 spikes.
Life Sci Alliance, 6, 2023
|
|
8H14
 
 | | Structure of SARS-CoV-1 Spike Protein with Engineered x3 Disulfide (D414C and V969C), Locked-1 Conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, LINOLEIC ACID, Spike glycoprotein | | Authors: | Zhang, X, Li, Z, Liu, Y, Wang, J, Fu, L, Wang, P, He, J, Xiong, X. | | Deposit date: | 2022-09-30 | | Release date: | 2022-10-19 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.39 Å) | | Cite: | Disulfide stabilization reveals conserved dynamic features between SARS-CoV-1 and SARS-CoV-2 spikes.
Life Sci Alliance, 6, 2023
|
|
8H10
 
 | | Structure of SARS-CoV-1 Spike Protein with Engineered x1 Disulfide (S370C and D967C), Locked-2 Conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BILIVERDINE IX ALPHA, ... | | Authors: | Zhang, X, Li, Z, Liu, Y, Wang, J, Fu, L, Wang, P, He, J, Xiong, X. | | Deposit date: | 2022-09-30 | | Release date: | 2022-10-19 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (2.99 Å) | | Cite: | Disulfide stabilization reveals conserved dynamic features between SARS-CoV-1 and SARS-CoV-2 spikes.
Life Sci Alliance, 6, 2023
|
|
8H16
 
 | | Structure of SARS-CoV-1 Spike Protein (S/native) at pH 5.5, Open Conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Zhang, X, Li, Z, Liu, Y, Wang, J, Fu, L, Wang, P, He, J, Xiong, X. | | Deposit date: | 2022-09-30 | | Release date: | 2022-11-09 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.35534 Å) | | Cite: | Disulfide stabilization reveals conserved dynamic features between SARS-CoV-1 and SARS-CoV-2 spikes.
Life Sci Alliance, 6, 2023
|
|
8H11
 
 | | Structure of SARS-CoV-1 Spike Protein with Engineered x1 Disulfide (S370C and D967C), Closed Conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Zhang, X, Li, Z, Liu, Y, Wang, J, Fu, L, Wang, P, He, J, Xiong, X. | | Deposit date: | 2022-09-30 | | Release date: | 2022-11-09 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (2.72 Å) | | Cite: | Disulfide stabilization reveals conserved dynamic features between SARS-CoV-1 and SARS-CoV-2 spikes.
Life Sci Alliance, 6, 2023
|
|
7DV8
 
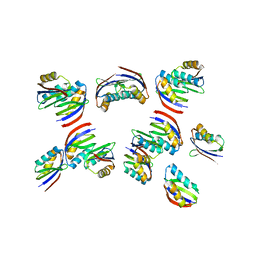 | | The crystal structure of rice immune receptor RGA5-HMA2. | | Descriptor: | Disease resistance protein RGA5 | | Authors: | Zhang, X, Liu, J.F. | | Deposit date: | 2021-01-12 | | Release date: | 2021-10-20 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.447 Å) | | Cite: | A designer rice NLR immune receptor confers resistance to the rice blast fungus carrying noncorresponding avirulence effectors.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
8H0Z
 
 | | Structure of SARS-CoV-1 Spike Protein with Engineered x1 Disulfide (S370C and D967C), Locked-122 Conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BILIVERDINE IX ALPHA, ... | | Authors: | Zhang, X, Li, Z, Liu, Y, Wang, J, Fu, L, Wang, P, He, J, Xiong, X. | | Deposit date: | 2022-09-30 | | Release date: | 2022-11-09 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.99 Å) | | Cite: | Disulfide stabilization reveals conserved dynamic features between SARS-CoV-1 and SARS-CoV-2 spikes.
Life Sci Alliance, 6, 2023
|
|
8H12
 
 | | Structure of SARS-CoV-1 Spike Protein with Engineered x2 Disulfide (G400C and V969C), Locked-2 Conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Zhang, X, Li, Z, Liu, Y, Wang, J, Fu, L, Wang, P, He, J, Xiong, X. | | Deposit date: | 2022-09-30 | | Release date: | 2022-11-09 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.44681 Å) | | Cite: | Disulfide stabilization reveals conserved dynamic features between SARS-CoV-1 and SARS-CoV-2 spikes.
Life Sci Alliance, 6, 2023
|
|
