7WS5
 
 | | Structures of Omicron Spike complexes illuminate broad-spectrum neutralizing antibody development | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 510A5 heavy chain, ... | | Authors: | Guo, H, Gao, Y, Ji, X, Yang, H. | | Deposit date: | 2022-01-28 | | Release date: | 2022-06-01 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structures of Omicron spike complexes and implications for neutralizing antibody development.
Cell Rep, 39, 2022
|
|
7WS1
 
 | | Structures of Omicron Spike complexes illuminate broad-spectrum neutralizing antibody development | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 510A5 heavy chain, ... | | Authors: | Guo, H, Gao, Y, Ji, X, Yang, H. | | Deposit date: | 2022-01-28 | | Release date: | 2022-06-01 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structures of Omicron spike complexes and implications for neutralizing antibody development.
Cell Rep, 39, 2022
|
|
7WS2
 
 | | Structures of Omicron Spike complexes illuminate broad-spectrum neutralizing antibody development | | Descriptor: | 510A5 heavy chain, 510A5 light chain, Spike protein S1 | | Authors: | Guo, H, Gao, Y, Ji, X, Yang, H. | | Deposit date: | 2022-01-28 | | Release date: | 2022-06-01 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structures of Omicron spike complexes and implications for neutralizing antibody development.
Cell Rep, 39, 2022
|
|
7WS7
 
 | | Structures of Omicron Spike complexes illuminate broad-spectrum neutralizing antibody development | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 510A5 heavy chain, 510A5 light chain, ... | | Authors: | Guo, H, Gao, Y, Ji, X, Yang, H. | | Deposit date: | 2022-01-28 | | Release date: | 2022-06-01 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structures of Omicron spike complexes and implications for neutralizing antibody development.
Cell Rep, 39, 2022
|
|
7E3L
 
 | | Ultrapotent SARS-CoV-2 neutralizing antibodies with protective efficacy against newly emerged mutational variants | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 58G6 heavy chain, 58G6 light chain, ... | | Authors: | Guo, H, Li, T, Liu, F, Gao, Y, Ji, X, Yang, H. | | Deposit date: | 2021-02-09 | | Release date: | 2021-09-15 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Potent SARS-CoV-2 neutralizing antibodies with protective efficacy against newly emerged mutational variants.
Nat Commun, 12, 2021
|
|
7E3K
 
 | | Ultrapotent SARS-CoV-2 neutralizing antibodies with protective efficacy against newly emerged mutational variants | | Descriptor: | 13G9 heavy chain, 13G9 light chain, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Guo, H, Li, T, Liu, F, Gao, Y, Ji, X, Yang, H. | | Deposit date: | 2021-02-09 | | Release date: | 2021-09-15 | | Last modified: | 2022-03-02 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Potent SARS-CoV-2 neutralizing antibodies with protective efficacy against newly emerged mutational variants.
Nat Commun, 12, 2021
|
|
5J5E
 
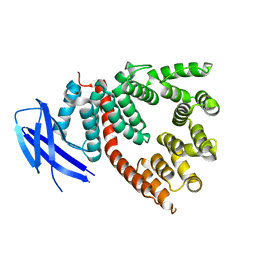 | | crystal structure of antigen-ERAP1 domain complex | | Descriptor: | Endoplasmic reticulum aminopeptidase 1 | | Authors: | Sui, L, Gandhi, A, Guo, H.-C. | | Deposit date: | 2016-04-02 | | Release date: | 2017-04-12 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of a polypeptide's C-terminus in complex with the regulatory domain of ER aminopeptidase 1.
Mol.Immunol., 80, 2016
|
|
2A3T
 
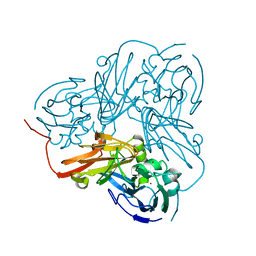 | | Cu-containing nitrite reductase | | Descriptor: | COPPER (II) ION, Copper-containing nitrite reductase, MAGNESIUM ION | | Authors: | Jacobson, F, Guo, H, Olesen, K, Okvist, M, Neutze, R, Sjolin, L. | | Deposit date: | 2005-06-27 | | Release date: | 2005-08-02 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structures of the oxidized and reduced forms of nitrite reductase from Rhodobacter sphaeroides 2.4.3 at high pH: changes in the interactions of the type 2 copper.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
2Q5X
 
 | |
6QLZ
 
 | | IDOL F3ab subdomain | | Descriptor: | E3 ubiquitin-protein ligase MYLIP | | Authors: | Martinelli, L, Johansson, P, Wan, P.T, Gunnarsson, J, Guo, H, Boyd, H. | | Deposit date: | 2019-02-01 | | Release date: | 2020-02-19 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.343 Å) | | Cite: | Structural analysis of the LDL receptor-interacting FERM domain in the E3 ubiquitin ligase IDOL reveals an obscured substrate-binding site.
J.Biol.Chem., 295, 2020
|
|
7OAR
 
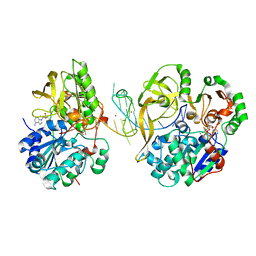 | | Crystal structure of helicase Pif1 from Thermus oshimai in complex with parallel G-quadruplex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DNA (28-MER), MAGNESIUM ION, ... | | Authors: | Dai, Y.X, Liu, N.N, Guo, H.L, Chen, W.F, Rety, S, Xi, X.G. | | Deposit date: | 2021-04-20 | | Release date: | 2022-03-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | Structural mechanism underpinning Thermus oshimai Pif1-mediated G-quadruplex unfolding.
Embo Rep., 23, 2022
|
|
5N98
 
 | | Crystal Structure of Drosophila DHX36 helicase in complex with TAGGGTTTT | | Descriptor: | CG9323, isoform A, DNA (5'-D(P*TP*AP*GP*GP*GP*TP*TP*TP*T)-3'), ... | | Authors: | Chen, W.-F, Rety, S, Guo, H.-L, Wu, W.-Q, Liu, N.-N, Liu, Q.-W, Dai, Y.-X, Xi, X.-G. | | Deposit date: | 2017-02-24 | | Release date: | 2018-03-14 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.756 Å) | | Cite: | Molecular Mechanistic Insights into Drosophila DHX36-Mediated G-Quadruplex Unfolding: A Structure-Based Model.
Structure, 26, 2018
|
|
5N94
 
 | | Crystal Structure of Drosophila DHX36 helicase in complex with polyU | | Descriptor: | CG9323, isoform A, RNA (5'-R(P*UP*UP*UP*UP*UP*UP*UP*U)-3') | | Authors: | Chen, W.-F, Rety, S, Hai-Lei Guo, H.-L, Wu, W.-Q, Liu, N.-N, Liu, Q.-W, Dai, Y.-X, Xi, X.-G. | | Deposit date: | 2017-02-24 | | Release date: | 2018-03-14 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.428 Å) | | Cite: | Molecular Mechanistic Insights into Drosophila DHX36-Mediated G-Quadruplex Unfolding: A Structure-Based Model.
Structure, 26, 2018
|
|
5N9D
 
 | | Crystal Structure of Drosophila DHX36 helicase in complex with GGGTTAGGGT | | Descriptor: | CG9323, isoform A, DNA (5'-D(P*GP*GP*GP*TP*TP*AP*GP*GP*GP*T)-3') | | Authors: | Chen, W.-F, Rety, S, Guo, H.-L, Wu, W.-Q, Liu, N.-N, Liu, Q.-W, Dai, Y.-X, Xi, X.-G. | | Deposit date: | 2017-02-24 | | Release date: | 2018-03-14 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | Molecular Mechanistic Insights into Drosophila DHX36-Mediated G-Quadruplex Unfolding: A Structure-Based Model.
Structure, 26, 2018
|
|
5V2I
 
 | |
5N96
 
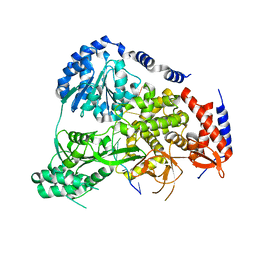 | | Crystal Structure of Drosophila DHX36 helicase in complex with AGGGTTTTTT | | Descriptor: | CG9323, isoform A, DNA (5'-D(P*AP*GP*GP*GP*TP*TP*TP*TP*TP*T)-3'), ... | | Authors: | Chen, W.-F, Rety, S, Guo, H.-L, Wu, W.-Q, Liu, N.-N, Liu, Q.-W, Dai, Y.-X, Xi, X.-G. | | Deposit date: | 2017-02-24 | | Release date: | 2018-03-14 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.716 Å) | | Cite: | Molecular Mechanistic Insights into Drosophila DHX36-Mediated G-Quadruplex Unfolding: A Structure-Based Model.
Structure, 26, 2018
|
|
5N8R
 
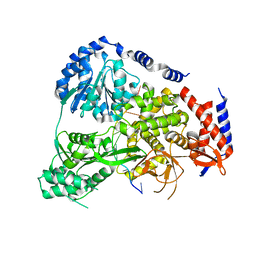 | | Crystal Structure of Drosophilia DHX36 helicase in complex with GAGCACTGC | | Descriptor: | CG9323, isoform A, DNA (5'-D(P*GP*AP*GP*CP*AP*CP*TP*GP*C)-3') | | Authors: | Chen, W.-F, Rety, S, Hai-Lei Guo, H.-L, Wu, W.-Q, Liu, N.-N, Liu, Q.-W, Dai, Y.-X, Xi, X.-G. | | Deposit date: | 2017-02-24 | | Release date: | 2018-03-14 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Molecular Mechanistic Insights into Drosophila DHX36-Mediated G-Quadruplex Unfolding: A Structure-Based Model.
Structure, 26, 2018
|
|
5N9A
 
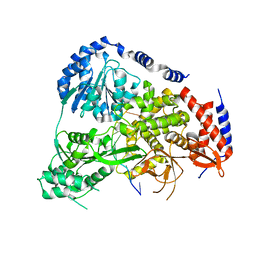 | | Crystal Structure of Drosophila DHX36 helicase in complex with GTTAGGGTT | | Descriptor: | CG9323, isoform A, DNA (5'-D(P*GP*TP*TP*AP*GP*GP*GP*TP*T)-3') | | Authors: | Chen, W.-F, Rety, S, Guo, H.-L, Wu, W.-Q, Liu, N.-N, Liu, Q.-W, Dai, Y.-X, Xi, X.-G. | | Deposit date: | 2017-02-24 | | Release date: | 2018-03-14 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.036 Å) | | Cite: | Molecular Mechanistic Insights into Drosophila DHX36-Mediated G-Quadruplex Unfolding: A Structure-Based Model.
Structure, 26, 2018
|
|
5N90
 
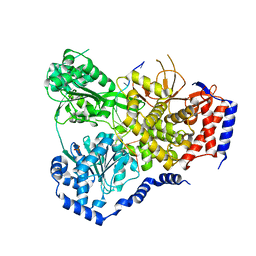 | | Crystal Structure of Drosophila DHX36 helicase in complex with TTGTGGTGT | | Descriptor: | CG9323, isoform A, DNA (5'-D(P*TP*TP*GP*TP*GP*GP*TP*GP*T)-3'), ... | | Authors: | Chen, W.-F, Rety, S, Guo, H.-L, Wu, W.-Q, Liu, N.-N, Liu, Q.-W, Dai, Y.-X, Xi, X.-G. | | Deposit date: | 2017-02-24 | | Release date: | 2018-03-14 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.069 Å) | | Cite: | Molecular Mechanistic Insights into Drosophila DHX36-Mediated G-Quadruplex Unfolding: A Structure-Based Model.
Structure, 26, 2018
|
|
5N8S
 
 | | Crystal Structure of Drosophila DHX36 helicase in complex with polyT | | Descriptor: | CG9323, isoform A, DNA (5'-D(P*TP*TP*TP*TP*TP*TP*TP*TP*TP*T)-3'), ... | | Authors: | Chen, W.-F, Rety, S, Hai-Lei Guo, H.-L, Wu, W.-Q, Liu, N.-N, Liu, Q.-W, Dai, Y.-X, Xi, X.-G. | | Deposit date: | 2017-02-24 | | Release date: | 2018-03-14 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.88 Å) | | Cite: | Molecular Mechanistic Insights into Drosophila DHX36-Mediated G-Quadruplex Unfolding: A Structure-Based Model.
Structure, 26, 2018
|
|
1EZL
 
 | | CRYSTAL STRUCTURE OF THE DISULPHIDE BOND-DEFICIENT AZURIN MUTANT C3A/C26A: HOW IMPORTANT IS THE S-S BOND FOR FOLDING AND STABILITY? | | Descriptor: | AZURIN, COPPER (II) ION | | Authors: | Bonander, N, Leckner, J, Guo, H, Karlsson, B.G, Sjolin, L. | | Deposit date: | 2000-05-11 | | Release date: | 2000-08-09 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the disulfide bond-deficient azurin mutant C3A/C26A: how important is the S-S bond for folding and stability?
Eur.J.Biochem., 267, 2000
|
|
5N8U
 
 | | Crystal Structure of Drosophila DHX36 helicase in complex with CTCTCCT | | Descriptor: | CG9323, isoform A, DNA (5'-D(P*CP*TP*CP*TP*CP*CP*CP*T)-3'), ... | | Authors: | Chen, W.-F, Rety, S, Guo, H.-L, Wu, W.-Q, Liu, N.-N, Liu, Q.-W, Dai, Y.-X, Xi, X.-G. | | Deposit date: | 2017-02-24 | | Release date: | 2018-03-14 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | Structural and mechanistic insights into DHX36-mediated innate immunity and G-quadruplex unfolding
To Be Published
|
|
5N9E
 
 | | Crystal Structure of Drosophila DHX36 helicase in complex with TGGGGATTT | | Descriptor: | CG9323, isoform A, DNA (5'-D(P*TP*GP*GP*GP*GP*AP*TP*TP*T)-3') | | Authors: | Chen, W.-F, Rety, S, Hai-Lei Guo, H.-L, Wu, W.-Q, Liu, N.-N, Liu, Q.-W, Dai, Y.-X, Xi, X.-G. | | Deposit date: | 2017-02-24 | | Release date: | 2018-03-21 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.007 Å) | | Cite: | Structural and mechanistic insights into DHX36-mediated innate immunity and G-quadruplex unfolding
To Be Published
|
|
5N8Z
 
 | | Crystal Structure of Drosophila DHX36 helicase in complex with CTCTCCCTT | | Descriptor: | CG9323, isoform A, DNA (5'-D(P*CP*TP*CP*TP*CP*CP*CP*TP*T)-3'), ... | | Authors: | Chen, W.-F, Rety, S, Guo, H.-L, Wu, W.-Q, Liu, N.-N, Liu, Q.-W, Dai, Y.-X, Xi, X.-G. | | Deposit date: | 2017-02-24 | | Release date: | 2018-03-14 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.477 Å) | | Cite: | Structural and mechanistic insights into DHX36-mediated innate immunity and G-quadruplex unfolding
To Be Published
|
|
5N9F
 
 | | Crystal Structure of Drosophila DHX36 helicase in complex with ssDNA CpG_A | | Descriptor: | CG9323, isoform A, CHLORIDE ION, ... | | Authors: | Chen, W.-F, Rety, S, Hai-Lei Guo, H.-L, Wu, W.-Q, Liu, N.-N, Liu, Q.-W, Dai, Y.-X, Xi, X.-G. | | Deposit date: | 2017-02-24 | | Release date: | 2018-03-21 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.969 Å) | | Cite: | Structural and mechanistic insights into DHX36-mediated innate immunity and G-quadruplex unfolding
To Be Published
|
|
