3LU1
 
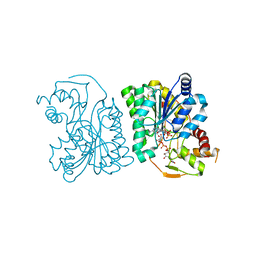 | | Crystal Structure Analysis of WbgU: a UDP-GalNAc 4-epimerase | | Descriptor: | GLYCINE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, SODIUM ION, ... | | Authors: | Bhatt, V.S, Guo, C.Y, Zhao, G, Yi, W, Liu, Z.J, Wang, P.G. | | Deposit date: | 2010-02-16 | | Release date: | 2010-07-21 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Altered architecture of substrate binding region defines the unique specificity of UDP-GalNAc 4-epimerases.
Protein Sci., 20, 2011
|
|
8I0H
 
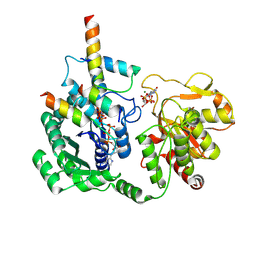 | |
7EDK
 
 | |
7WXF
 
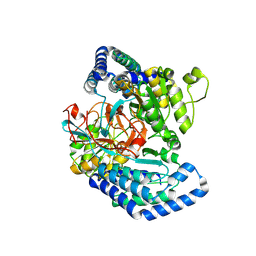 | |
7WXH
 
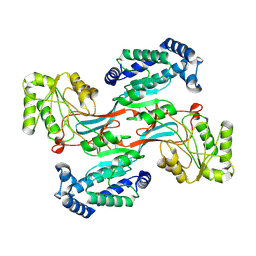 | | GPR domain open form of Drosophila P5CS filament with glutamate, ATP, and NADPH | | Descriptor: | Delta-1-pyrroline-5-carboxylate synthase | | Authors: | Liu, J.L, Zhong, J, Guo, C.J, Zhou, X. | | Deposit date: | 2022-02-14 | | Release date: | 2022-03-30 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Structural basis of dynamic P5CS filaments.
Elife, 11, 2022
|
|
7WXG
 
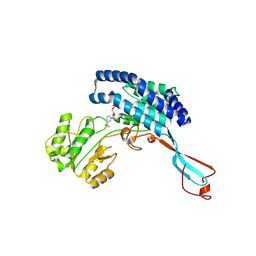 | | GPR domain closed form of Drosophila P5CS filament with glutamate, ATP, and NADPH | | Descriptor: | Delta-1-pyrroline-5-carboxylate synthase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Liu, J.L, Zhong, J, Guo, C.J, Zhou, X. | | Deposit date: | 2022-02-14 | | Release date: | 2022-03-30 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Structural basis of dynamic P5CS filaments.
Elife, 11, 2022
|
|
7WX4
 
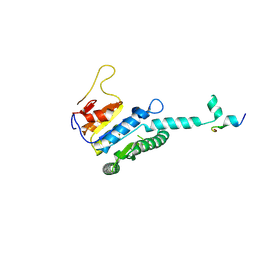 | |
7FJD
 
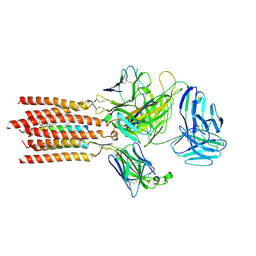 | | Cryo-EM structure of a membrane protein(WT) | | Descriptor: | CHOLESTEROL, T cell receptor alpha variable 12-3,Possible J 11 gene segment,T cell receptor alpha chain constant, T cell receptor beta variable 6-5,M1-specific T cell receptor beta chain,T cell receptor beta constant 2, ... | | Authors: | Chen, Y, Zhu, Y, Gao, W, Zhang, A, Guo, C, Huang, Z. | | Deposit date: | 2021-08-03 | | Release date: | 2022-07-27 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Cholesterol inhibits TCR signaling by directly restricting TCR-CD3 core tunnel motility.
Mol.Cell, 82, 2022
|
|
7FJE
 
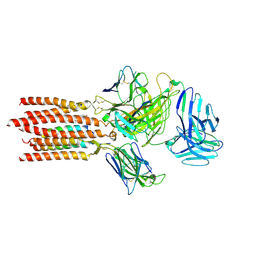 | | Cryo-EM structure of a membrane protein(LL) | | Descriptor: | CHOLESTEROL, T cell receptor alpha variable 12-3,Possible J 11 gene segment,T cell receptor alpha chain constant, T cell receptor beta variable 6-5,M1-specific T cell receptor beta chain,T cell receptor beta constant 2, ... | | Authors: | Chen, Y, Zhu, Y, Gao, W, Zhang, A, Guo, C, Huang, Z. | | Deposit date: | 2021-08-03 | | Release date: | 2022-07-27 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Cholesterol inhibits TCR signaling by directly restricting TCR-CD3 core tunnel motility.
Mol.Cell, 82, 2022
|
|
7FJF
 
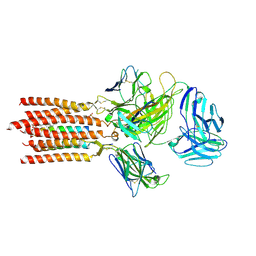 | | Cryo-EM structure of a membrane protein(CS) | | Descriptor: | CHOLEST-5-EN-3-YL HYDROGEN SULFATE, T cell receptor alpha variable 12-3,Possible J 11 gene segment,T cell receptor alpha chain constant, T cell receptor beta variable 6-5,M1-specific T cell receptor beta chain,T cell receptor beta constant 2, ... | | Authors: | Chen, Y, Zhu, Y, Gao, W, Zhang, A, Guo, C, Huang, Z. | | Deposit date: | 2021-08-03 | | Release date: | 2022-07-27 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Cholesterol inhibits TCR signaling by directly restricting TCR-CD3 core tunnel motility.
Mol.Cell, 82, 2022
|
|
7WXI
 
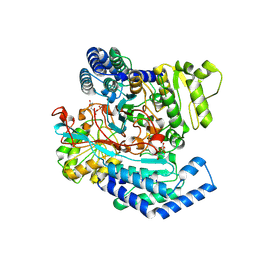 | | GPR domain of Drosophila P5CS filament with glutamate and ATPgammaS | | Descriptor: | Delta-1-pyrroline-5-carboxylate synthase, GAMMA-GLUTAMYL PHOSPHATE | | Authors: | Liu, J.L, Zhong, J, Guo, C.J, Zhou, X. | | Deposit date: | 2022-02-14 | | Release date: | 2022-03-30 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Structural basis of dynamic P5CS filaments.
Elife, 11, 2022
|
|
7WX3
 
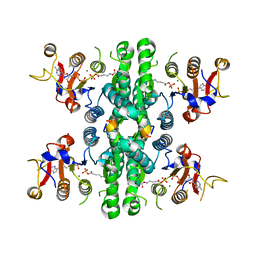 | | GK domain of Drosophila P5CS filament with glutamate, ATP, and NADPH | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Delta-1-pyrroline-5-carboxylate synthase, GAMMA-GLUTAMYL PHOSPHATE, ... | | Authors: | Liu, J.L, Zhong, J, Guo, C.J, Zhou, X. | | Deposit date: | 2022-02-14 | | Release date: | 2022-04-06 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural basis of dynamic P5CS filaments.
Elife, 11, 2022
|
|
7WJ4
 
 | |
7WIZ
 
 | | Structural basis for ligand binding modes of CTP synthase | | Descriptor: | CTP synthase, GLUTAMINE, PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER, ... | | Authors: | Liu, J.L, Guo, C.J. | | Deposit date: | 2022-01-05 | | Release date: | 2023-01-11 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural basis for ligand binding modes of CTP synthase.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
5YMR
 
 | | The Crystal Structure of IseG | | Descriptor: | 2-hydroxyethylsulfonic acid, Formate acetyltransferase, GLYCEROL | | Authors: | Lin, L, Zhang, J, Xing, M, Hua, G, Guo, C, Hu, Y, Wei, Y, Ang, E, Zhao, H, Zhang, Y, Yuchi, Z. | | Deposit date: | 2017-10-22 | | Release date: | 2019-03-20 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Radical-mediated C-S bond cleavage in C2 sulfonate degradation by anaerobic bacteria.
Nat Commun, 10, 2019
|
|
7F5V
 
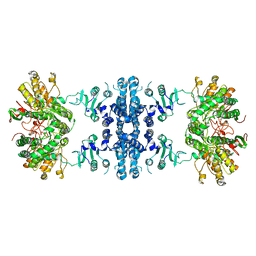 | | Drosophila P5CS filament with glutamate, ATP, and NADPH | | Descriptor: | Delta-1-pyrroline-5-carboxylate synthase | | Authors: | Liu, J.L, Zhong, J, Guo, C.J, Zhou, X. | | Deposit date: | 2021-06-22 | | Release date: | 2022-04-06 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural basis of dynamic P5CS filaments.
Elife, 11, 2022
|
|
7F5U
 
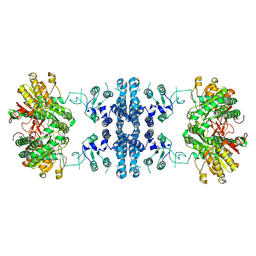 | |
7F5X
 
 | | GK domain of Drosophila P5CS filament with glutamate | | Descriptor: | Delta-1-pyrroline-5-carboxylate synthase, GAMMA-L-GLUTAMIC ACID | | Authors: | Liu, J.L, Zhong, J, Guo, C.J, Zhou, X. | | Deposit date: | 2021-06-23 | | Release date: | 2022-04-06 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural basis of dynamic P5CS filaments.
Elife, 11, 2022
|
|
7F5T
 
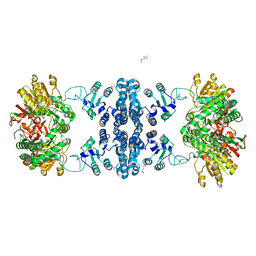 | | Drosophila P5CS filament with glutamate | | Descriptor: | Delta-1-pyrroline-5-carboxylate synthase, GLUTAMIC ACID | | Authors: | Liu, J.L, Zhong, J, Guo, C.J, Zhou, X. | | Deposit date: | 2021-06-22 | | Release date: | 2022-05-18 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Structural basis of dynamic P5CS filaments.
Elife, 11, 2022
|
|
7DPT
 
 | | Structural basis for ligand binding modes of CTP synthase | | Descriptor: | 6-DIAZENYL-5-OXO-L-NORLEUCINE, ADENOSINE-5'-DIPHOSPHATE, CTP synthase, ... | | Authors: | Liu, J.L, Zhou, X, Guo, C.J, Chang, C.C. | | Deposit date: | 2020-12-21 | | Release date: | 2021-09-15 | | Method: | ELECTRON MICROSCOPY (2.48 Å) | | Cite: | Structural basis for ligand binding modes of CTP synthase.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7DPW
 
 | | Structural basis for ligand binding modes of CTP synthase | | Descriptor: | CTP synthase, CYTIDINE-5'-TRIPHOSPHATE, MAGNESIUM ION | | Authors: | Liu, J.L, Zhou, X, Guo, C.J, Chang, C.C. | | Deposit date: | 2020-12-21 | | Release date: | 2021-09-15 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (2.65 Å) | | Cite: | Structural basis for ligand binding modes of CTP synthase.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7YMD
 
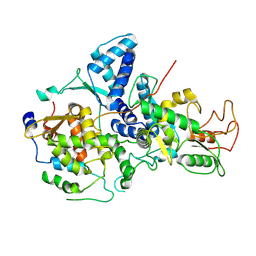 | | Cryo-EM structure of Nse1/3/4 | | Descriptor: | Non-structural maintenance of chromosome element 3, Non-structural maintenance of chromosome element 4, Non-structural maintenance of chromosomes element 1 | | Authors: | Qian, L, Jun, Z, Zhenguo, C, Wang, L. | | Deposit date: | 2022-07-28 | | Release date: | 2024-01-31 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (4.176 Å) | | Cite: | Cryo-EM structures of Smc5/6 in multiple states reveal its assembly and functional mechanisms.
Nat.Struct.Mol.Biol., 2024
|
|
8HQY
 
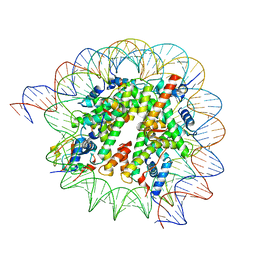 | | Cryo-EM structure of SSX1 bound to the H2AK119Ub nucleosome at a resolution of 3.05 angstrom | | Descriptor: | DNA (136-MER), DNA (137-MER), Histone H2A type 1-B/E, ... | | Authors: | Zebin, T, Ai, H.S, Ziyu, X, GuoChao, C, Man, P, Liu, L. | | Deposit date: | 2022-12-14 | | Release date: | 2023-09-27 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.05 Å) | | Cite: | Synovial sarcoma X breakpoint 1 protein uses a cryptic groove to selectively recognize H2AK119Ub nucleosomes.
Nat.Struct.Mol.Biol., 31, 2024
|
|
7YLM
 
 | | Cryo-EM structure of 8-subunit Smc5/6 hinge region | | Descriptor: | MMS21 isoform 1, SMC6 isoform 1, Structural maintenance of chromosomes protein 5 | | Authors: | Qian, L, Jun, Z, Xiang, Z, Wang, Z, Tong, C, Duo, J, Zhenguo, C, Wang, L. | | Deposit date: | 2022-07-26 | | Release date: | 2024-01-31 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (6.17 Å) | | Cite: | Cryo-EM structures of Smc5/6 in multiple states reveal its assembly and functional mechanisms.
Nat.Struct.Mol.Biol., 2024
|
|
7YQH
 
 | | Cryo-EM structure of 8-subunit Smc5/6 | | Descriptor: | DNA repair protein KRE29, E3 SUMO-protein ligase MMS21, Non-structural maintenance of chromosome element 3, ... | | Authors: | Qian, L, Jun, Z, Xiang, Z, Tong, C, Zhaoning, W, Duo, J, Zhenguo, C, Lanfeng, W. | | Deposit date: | 2022-08-07 | | Release date: | 2024-01-31 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (5.6 Å) | | Cite: | Cryo-EM structures of Smc5/6 in multiple states reveal its assembly and functional mechanisms.
Nat.Struct.Mol.Biol., 2024
|
|
