1BUS
 
 | |
2HOA
 
 | | STRUCTURE DETERMINATION OF THE ANTP(C39->S) HOMEODOMAIN FROM NUCLEAR MAGNETIC RESONANCE DATA IN SOLUTION USING A NOVEL STRATEGY FOR THE STRUCTURE CALCULATION WITH THE PROGRAMS DIANA, CALIBA, HABAS AND GLOMSA | | Descriptor: | ANTENNAPEDIA PROTEIN | | Authors: | Guntert, P, Qian, Y.-Q, Otting, G, Muller, M, Gehring, W.J, Wuthrich, K. | | Deposit date: | 1992-04-04 | | Release date: | 1993-10-31 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structure determination of the Antp (C39----S) homeodomain from nuclear magnetic resonance data in solution using a novel strategy for the structure calculation with the programs DIANA, CALIBA, HABAS and GLOMSA.
J.Mol.Biol., 217, 1991
|
|
2BUS
 
 | |
1VDY
 
 | | NMR Structure of the hypothetical ENTH-VHS domain At3g16270 from Arabidopsis thaliana | | Descriptor: | hypothetical protein (RAFL09-17-B18) | | Authors: | Lopez-Mendez, B, Pantoja-Uceda, D, Tomizawa, T, Koshiba, S, Kigawa, T, Shirouzu, M, Terada, T, Inoue, M, Yabuki, T, Aoki, M, Seki, E, Matsuda, T, Hirota, H, Yoshida, M, Tanaka, A, Osanai, T, Seki, M, Shinozaki, K, Yokoyama, S, Guntert, P, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-03-25 | | Release date: | 2005-05-03 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the hypothetical ENTH-VHS domain AT3G16270 from arabidopsis thaliana
To be Published
|
|
8PXS
 
 | | Short RNA binding to peptide amyloids | | Descriptor: | RNA (5'-R(P*GP*UP*CP*A)-3'), VAL-ALA-GLN-ALA-GLN-ILE-ASN-ILE | | Authors: | Rout, S.K, Cadalbert, R, Schroder, N, Wiegand, T, Zehnder, J, Gampp, O, Guntert, P, Kringler, D, Kreutz, C, Knorlein, A, Hall, J, Greenwald, J, Riek, R. | | Deposit date: | 2023-07-24 | | Release date: | 2023-10-18 | | Last modified: | 2024-10-16 | | Method: | SOLID-STATE NMR | | Cite: | An Analysis of Nucleotide-Amyloid Interactions Reveals Selective Binding to Codon-Sized RNA.
J.Am.Chem.Soc., 145, 2023
|
|
5GVQ
 
 | | Solution structure of the first RRM domain of human spliceosomal protein SF3b49 | | Descriptor: | Splicing factor 3B subunit 4 | | Authors: | Kuwasako, K, Nameki, N, Tsuda, K, Takahashi, M, Sato, A, Tochio, N, Inoue, M, Terada, T, Kigawa, T, Kobayashi, N, Shirouzu, M, Ito, T, Sakamoto, T, Wakamatsu, K, Guntert, P, Takahashi, S, Yokoyama, S, Muto, Y, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2016-09-06 | | Release date: | 2017-04-12 | | Last modified: | 2025-03-19 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the first RNA recognition motif domain of human spliceosomal protein SF3b49 and its mode of interaction with a SF3b145 fragment.
Protein Sci., 26, 2017
|
|
7QTR
 
 | | GB1 in mammalian cells, 50 uM | | Descriptor: | Immunoglobulin G-binding protein G | | Authors: | Gerez, J.A, Prymaczok, N.C, Kadavath, H, Gosh, D, Butikofer, M, Guntert, P, Riek, R. | | Deposit date: | 2022-01-15 | | Release date: | 2022-12-21 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Protein structure determination in human cells by in-cell NMR and a reporter system to optimize protein delivery or transexpression.
Commun Biol, 5, 2022
|
|
7QTS
 
 | | GB1 in mammalian cells, 10 uM | | Descriptor: | Immunoglobulin G-binding protein G | | Authors: | Gerez, J.A, Prymaczok, N.C, Kadavath, H, Gosh, D, Butikofer, M, Guntert, P, Riek, R. | | Deposit date: | 2022-01-15 | | Release date: | 2022-12-21 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Protein structure determination in human cells by in-cell NMR and a reporter system to optimize protein delivery or transexpression.
Commun Biol, 5, 2022
|
|
6FGN
 
 | | Solution Structure of p300Taz2-p63TA | | Descriptor: | Histone acetyltransferase p300,Tumor protein 63, ZINC ION | | Authors: | Gebel, J, Kazemi, S, Lohr, F, Guntert, P, Dotsch, V. | | Deposit date: | 2018-01-11 | | Release date: | 2018-05-30 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Regulation of the Activity in the p53 Family Depends on the Organization of the Transactivation Domain.
Structure, 26, 2018
|
|
8Y4Z
 
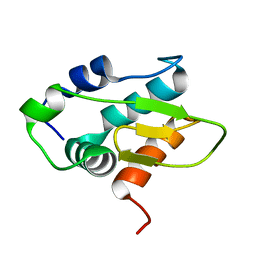 | | Monomeric HERC5 HECT c-lobe structure in solution | | Descriptor: | E3 ISG15--protein ligase HERC5 | | Authors: | Dag, C, Lambert, M, Kahraman, K, Lohn, F, Lee, W, Gocenler, O, Guntert, P, Dotsch, V. | | Deposit date: | 2024-01-31 | | Release date: | 2024-02-14 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Monomeric HERC5 HECT c-lobe structure in solution
To Be Published
|
|
1GM0
 
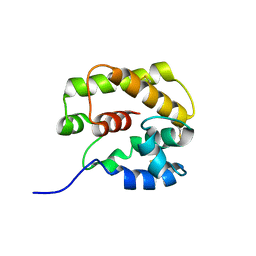 | | A Form of the Pheromone-Binding Protein from Bombyx mori | | Descriptor: | PHEROMONE-BINDING PROTEIN | | Authors: | Horst, R, Damberger, F, Guntert, P, Luginbuhl, P, Nikonova, L, Peng, G, Leal, W.S, Wuthrich, K. | | Deposit date: | 2001-09-05 | | Release date: | 2001-11-30 | | Last modified: | 2024-10-23 | | Method: | SOLUTION NMR | | Cite: | NMR Structure Reveals Novel Intramolecular Regulation Mechanism for Pheromone-Binding and Release
Proc.Natl.Acad.Sci.USA, 98, 2001
|
|
7X9U
 
 | | Type-II KH motif of human mitochondrial RbfA | | Descriptor: | Putative ribosome-binding factor A, mitochondrial | | Authors: | Kuwasako, K, Suzuki, S, Furue, M, Takizawa, M, Takahashi, M, Tsuda, K, Nagata, T, Watanabe, S, Tanaka, A, Kobayashi, N, Kigawa, T, Guntert, P, Shirouzu, M, Yokoyama, S, Muto, Y, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2022-03-16 | | Release date: | 2023-01-25 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | 1 H, 13 C, and 15 N resonance assignments and solution structures of the KH domain of human ribosome binding factor A, mtRbfA, involved in mitochondrial ribosome biogenesis.
Biomol.Nmr Assign., 16, 2022
|
|
1HIC
 
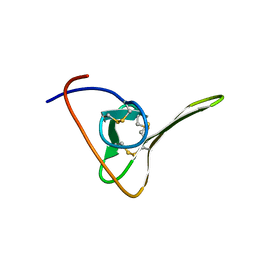 | |
1LG4
 
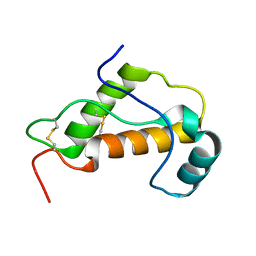 | |
5N6R
 
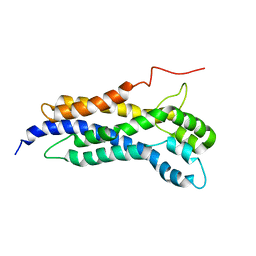 | | Solution structure of the Dbl-homology domain of Bcr-Abl | | Descriptor: | Breakpoint cluster region protein | | Authors: | Reckel, S, Lohr, F, Buchner, L, Guntert, P, Dotsch, V, Hantschel, O. | | Deposit date: | 2017-02-16 | | Release date: | 2017-12-27 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Structural and functional dissection of the DH and PH domains of oncogenic Bcr-Abl tyrosine kinase.
Nat Commun, 8, 2017
|
|
1HHN
 
 | | Calreticulin P-domain | | Descriptor: | CALRETICULIN | | Authors: | Ellgaard, L, Riek, R, Herrmann, T, Guntert, P, Braun, D, Helenius, A, Wuthrich, K. | | Deposit date: | 2000-12-22 | | Release date: | 2001-03-08 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | NMR Structure of the Calreticulin P-Domain
Proc.Natl.Acad.Sci.USA, 98, 2001
|
|
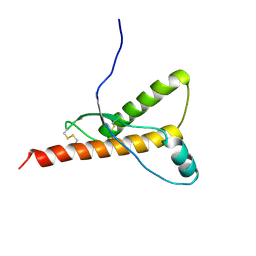
1H0L
 
 | |
1VD0
 
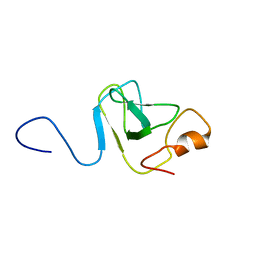 | | Capsid stabilizing protein GPD, NMR, 20 Structures | | Descriptor: | Head decoration protein | | Authors: | Iwai, H, Forrer, P, Pluckthun, A, Guntert, P, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-03-17 | | Release date: | 2005-03-29 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | NMR solution structure of the monomeric form of the bacteriophage lambda capsid stabilizing protein gpD.
J.Biomol.Nmr, 31, 2005
|
|
6F98
 
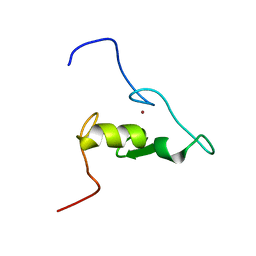 | | Solution structure of the RING domain of the E3 ubiquitin ligase HRD1 | | Descriptor: | E3 ubiquitin-protein ligase, ZINC ION | | Authors: | Kniss, A, Kazemi, S, Lohr, F, Guntert, P, Dotsch, V. | | Deposit date: | 2017-12-14 | | Release date: | 2019-01-30 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the RING domain of the E3 ubiquitin ligase HRD1
To Be Published
|
|
6SVH
 
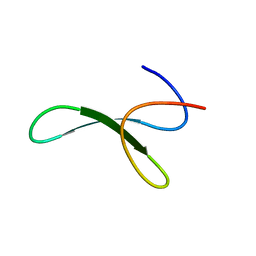 | | Protein allostery of the WW domain at atomic resolution: FFpSPR bound structure | | Descriptor: | Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1 | | Authors: | Strotz, D, Orts, J, Friedmann, M, Guntert, P, Vogeli, B, Riek, R. | | Deposit date: | 2019-09-18 | | Release date: | 2020-09-30 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Protein Allostery at Atomic Resolution.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
6SVE
 
 | | Protein allostery of the WW domain at atomic resolution: pCdc25C bound structure | | Descriptor: | Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1 | | Authors: | Strotz, D, Orts, J, Friedmann, M, Guntert, P, Vogeli, B, Riek, R. | | Deposit date: | 2019-09-18 | | Release date: | 2020-10-07 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Protein Allostery at Atomic Resolution.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
6FGS
 
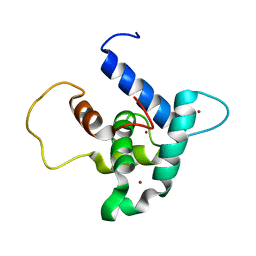 | | Solution structure of p300Taz2-p73TA1 | | Descriptor: | Histone acetyltransferase p300,Tumor protein p73, ZINC ION | | Authors: | Gebel, J, Kazemi, S, Lohr, F, Guntert, P, Dotsch, V. | | Deposit date: | 2018-01-11 | | Release date: | 2018-05-30 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Regulation of the Activity in the p53 Family Depends on the Organization of the Transactivation Domain.
Structure, 26, 2018
|
|
1K9C
 
 | | Solution Structure of Calreticulin P-domain subdomain (residues 189-261) | | Descriptor: | CALRETICULIN | | Authors: | Ellgaard, L, Bettendorff, P, Braun, D, Herrmann, T, Fiorito, F, Guntert, P, Helenius, A, Wuthrich, K. | | Deposit date: | 2001-10-29 | | Release date: | 2002-10-12 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR Structures of 36 and 73-residue Fragments of the Calreticulin P-domain
J.Mol.Biol., 322, 2002
|
|
1K91
 
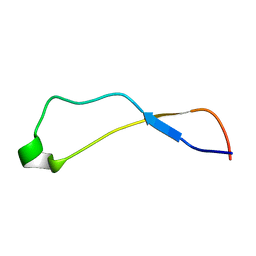 | | Solution Structure of Calreticulin P-domain subdomain (residues 221-256) | | Descriptor: | CALRETICULIN | | Authors: | Ellgaard, L, Bettendorff, P, Braun, D, Herrmann, T, Fiorito, F, Guntert, P, Helenius, A, Wuthrich, K. | | Deposit date: | 2001-10-26 | | Release date: | 2002-10-12 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR Structures of 36 and 73-residue Fragments of the Calreticulin P-domain
J.Mol.Biol., 322, 2002
|
|
1SZL
 
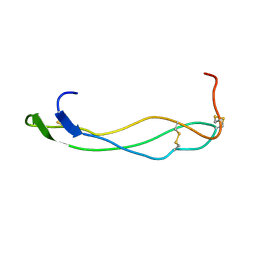 | | F-spondin TSR domain 1 | | Descriptor: | F-spondin | | Authors: | Tossavainen, H, Paakkonen, K, Permi, P, Kilpelainen, I, Guntert, P. | | Deposit date: | 2004-04-06 | | Release date: | 2005-04-19 | | Last modified: | 2024-11-06 | | Method: | SOLUTION NMR | | Cite: | Solution structures of the first and fourth TSR domains of F-spondin
Proteins, 64, 2006
|
|
