8PAY
 
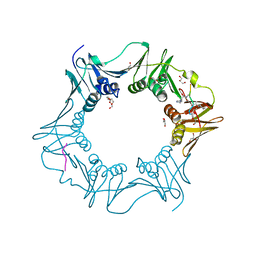 | | Structure of the E.coli DNA polymerase sliding clamp with a covalently bound peptide 2. | | Descriptor: | ACE-GLN-ALC-GLC-LEU-PHE, Beta sliding clamp, GLYCEROL, ... | | Authors: | Compain, G, Monsarrat, C, Blagojevic, J, Brillet, K, Dumas, P, Hammann, P, Kuhn, L, Martiel, I, Engilberge, S, Olieric, V, Wolff, P, Burnouf, D, wagner, J, Guichard, G. | | Deposit date: | 2023-06-08 | | Release date: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.21 Å) | | Cite: | Peptide-Based Covalent Inhibitors Bearing Mild Electrophiles to Target a Conserved His Residue of the Bacterial Sliding Clamp.
Jacs Au, 4, 2024
|
|
8PAT
 
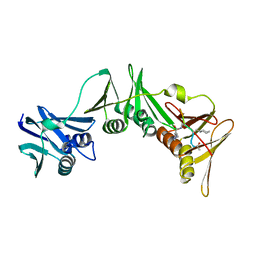 | | Structure of the E.coli DNA polymerase sliding clamp with a covalently bound peptide 3. | | Descriptor: | ACE-GLN-ALC-GLX-LEU-PHE, Beta sliding clamp | | Authors: | Compain, G, Monsarrat, C, Blagojevic, J, Brillet, K, Dumas, P, Hammann, P, Kuhn, L, Martiel, I, Engilberge, S, Olieric, V, Wolff, P, Burnouf, D, Guichard, G. | | Deposit date: | 2023-06-08 | | Release date: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Peptide-Based Covalent Inhibitors Bearing Mild Electrophiles to Target a Conserved His Residue of the Bacterial Sliding Clamp.
Jacs Au, 4, 2024
|
|
4ZER
 
 | | Crystal structure of the Onc112 antimicrobial peptide bound to the Thermus thermophilus 70S ribosome | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 16s ribosomal RNA, 23s ribosomal RNA, ... | | Authors: | Seefeldt, A.C, Nguyen, F, Antunes, S, Perebaskine, N, Graf, M, Arenz, S, Inampudi, K.K, Douat, C, Guichard, G, Wilson, D.N, Innis, C.A. | | Deposit date: | 2015-04-20 | | Release date: | 2015-05-20 | | Last modified: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | The proline-rich antimicrobial peptide Onc112 inhibits translation by blocking and destabilizing the initiation complex.
Nat.Struct.Mol.Biol., 22, 2015
|
|
8BV1
 
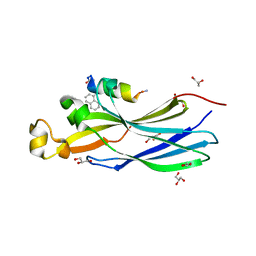 | | Peptide inhibitor P4 in complex with ASF1 histone chaperone | | Descriptor: | GLYCEROL, Histone chaperone ASF1A, P4 peptide inhibitor of histone chaperone ASF1 | | Authors: | Perrin, M.E, Li, B, Mbianda, J, Ropars, V, Legrand, P, Douat, C, Ochsenbein, F, Guichard, G. | | Deposit date: | 2022-12-01 | | Release date: | 2023-07-05 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.834 Å) | | Cite: | Unexpected binding modes of inhibitors to the histone chaperone ASF1 revealed by a foldamer scanning approach.
Chem.Commun.(Camb.), 59, 2023
|
|
8CJ3
 
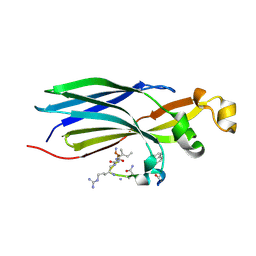 | | Urea-based foldamer inhibitor c3u_7 chimera in complex with ASF1 histone chaperone | | Descriptor: | Histone chaperone ASF1A, c3u_7 chimera inhibitor of histone chaperone ASF1 | | Authors: | Perrin, M.E, Li, B, Mbianda, J, Ropars, V, Legrand, P, Douat, C, Ochsenbein, F, Guichard, G. | | Deposit date: | 2023-02-11 | | Release date: | 2023-07-05 | | Last modified: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Unexpected binding modes of inhibitors to the histone chaperone ASF1 revealed by a foldamer scanning approach.
Chem.Commun.(Camb.), 59, 2023
|
|
8CJ1
 
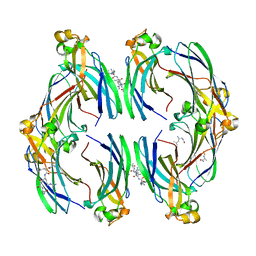 | | Urea-based foldamer inhibitor c3u_3 chimera in complex with ASF1 histone chaperone | | Descriptor: | Histone chaperone ASF1A, c3u_3 chimera inhibitor of histone chaperone ASF1 | | Authors: | Perrin, M.E, Li, B, Mbianda, J, Ropars, V, Legrand, P, Douat, C, Ochsenbein, F, Guichard, G. | | Deposit date: | 2023-02-11 | | Release date: | 2023-07-05 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.564 Å) | | Cite: | Unexpected binding modes of inhibitors to the histone chaperone ASF1 revealed by a foldamer scanning approach.
Chem.Commun.(Camb.), 59, 2023
|
|
8CJ2
 
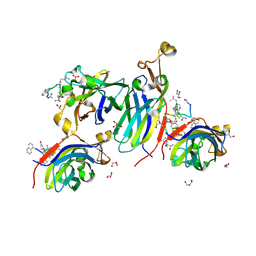 | | Urea-based foldamer inhibitor c3u_5 chimera in complex with ASF1 histone chaperone | | Descriptor: | GLYCEROL, Histone chaperone ASF1A, SULFATE ION, ... | | Authors: | Perrin, M.E, Li, B, Mbianda, J, Ropars, V, Legrand, P, Douat, C, Ochsenbein, F, Guichard, G. | | Deposit date: | 2023-02-11 | | Release date: | 2023-07-05 | | Last modified: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (2.127 Å) | | Cite: | Unexpected binding modes of inhibitors to the histone chaperone ASF1 revealed by a foldamer scanning approach.
Chem.Commun.(Camb.), 59, 2023
|
|
3Q4J
 
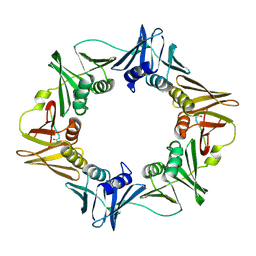 | | Structure of a small peptide ligand bound to E.coli DNA sliding clamp | | Descriptor: | DNA polymerase III subunit beta, peptide ligand | | Authors: | Wolff, P, Olieric, V, Briand, J.P, Chaloin, O, Dejaegere, A, Dumas, P, Ennifar, E, Guichard, G, Wagner, J, Burnouf, D. | | Deposit date: | 2010-12-23 | | Release date: | 2011-12-28 | | Last modified: | 2013-03-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure-based design of short peptide ligands binding onto the E. coli processivity ring.
J.Med.Chem., 54, 2011
|
|
3Q4L
 
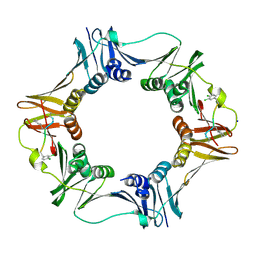 | | Structure of a small peptide ligand bound to E.coli DNA sliding clamp | | Descriptor: | DNA polymerase III subunit beta, SODIUM ION, peptide ligand | | Authors: | Wolff, P, Olieric, V, Briand, J.P, Chaloin, O, Dejaegere, A, Dumas, P, Ennifar, E, Guichard, G, Wagner, J, Burnouf, D. | | Deposit date: | 2010-12-23 | | Release date: | 2011-12-28 | | Last modified: | 2013-03-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure-based design of short peptide ligands binding onto the E. coli processivity ring.
J.Med.Chem., 54, 2011
|
|
3Q4K
 
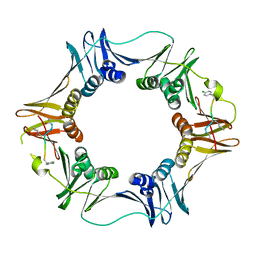 | | Structure of a small peptide ligand bound to E.coli DNA sliding clamp | | Descriptor: | DNA polymerase III subunit beta, peptide ligand | | Authors: | Wolff, P, Olieric, V, Briand, J.P, Chaloin, O, Dejaegere, A, Dumas, P, Ennifar, E, Guichard, G, Wagner, J, Burnouf, D. | | Deposit date: | 2010-12-23 | | Release date: | 2011-12-28 | | Last modified: | 2013-03-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure-based design of short peptide ligands binding onto the E. coli processivity ring.
J.Med.Chem., 54, 2011
|
|
1BFW
 
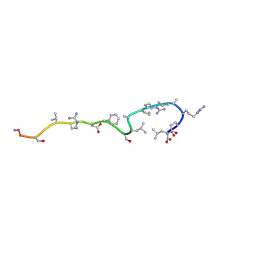 | | RETRO-INVERSO ANALOGUE OF THE G-H LOOP OF VP1 IN FOOT-AND-MOUTH-DISEASE (FMD) VIRUS, NMR, 10 STRUCTURES | | Descriptor: | VP1 PROTEIN | | Authors: | Petit, M.C, Benkirane, N, Guichard, G, Phan Chan Du, A, Cung, M.T, Briand, J.P, Muller, S. | | Deposit date: | 1998-05-22 | | Release date: | 1999-01-13 | | Last modified: | 2017-11-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a retro-inverso peptide analogue mimicking the foot-and-mouth disease virus major antigenic site. Structural basis for its antigenic cross-reactivity with the parent peptide.
J.Biol.Chem., 274, 1999
|
|
1BCV
 
 | | SYNTHETIC PEPTIDE CORRESPONDING TO THE MAJOR IMMUNOGEN SITE OF FMD VIRUS, NMR, 10 STRUCTURES | | Descriptor: | PEPTIDE CORRESPONDING TO THE MAJOR IMMUNOGEN SITE OF FMD VIRUS | | Authors: | Petit, M.C, Benkirane, N, Guichard, G, Phan Chan Du, A, Cung, M.T, Briand, J.P, Muller, S. | | Deposit date: | 1998-05-03 | | Release date: | 1998-11-25 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a retro-inverso peptide analogue mimicking the foot-and-mouth disease virus major antigenic site. Structural basis for its antigenic cross-reactivity with the parent peptide.
J.Biol.Chem., 274, 1999
|
|
1CVQ
 
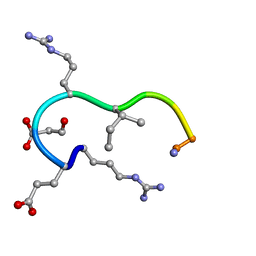 | | SOLUTION STRUCTURE OF THE ANALOGUE RETRO-INVERSO MGREGRIGGC IN CONTACT WITH THE MONOCLONAL ANTIBODY MAB 4X11, NMR, 7 STRUCTURES | | Descriptor: | HISTONE H3 | | Authors: | Phan Chan Du, A, Petit, M.C, Guichard, G, Briand, J.P, Muller, S, Cung, M.T. | | Deposit date: | 1999-08-24 | | Release date: | 1999-09-02 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | Structure of antibody-bound peptides and retro-inverso analogues. A transferred nuclear Overhauser effect spectroscopy and molecular dynamics approach.
Biochemistry, 40, 2001
|
|
6XZJ
 
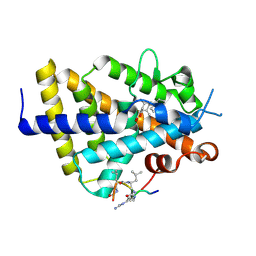 | | Structure of zVDR LBD-Calcitriol in complex with chimera 12 | | Descriptor: | 5-{2-[1-(5-HYDROXY-1,5-DIMETHYL-HEXYL)-7A-METHYL-OCTAHYDRO-INDEN-4-YLIDENE]-ETHYLIDENE}-4-METHYLENE-CYCLOHEXANE-1,3-DIOL, ACETATE ION, ARG-HIS-LYS-ILE-LEU-URR-UIL-URL-GLN, ... | | Authors: | Buratto, J, Belorusova, A.Y, Rochel, N, Guichard, G. | | Deposit date: | 2020-02-04 | | Release date: | 2021-02-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Basis for alpha-Helix Mimicry and Inhibition of Protein-Protein Interactions with Oligourea Foldamers.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
6XZH
 
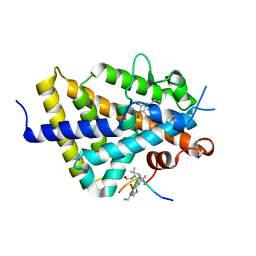 | | Structure of zVDR LBD-Calcitriol in complex with chimera 10 | | Descriptor: | 5-{2-[1-(5-HYDROXY-1,5-DIMETHYL-HEXYL)-7A-METHYL-OCTAHYDRO-INDEN-4-YLIDENE]-ETHYLIDENE}-4-METHYLENE-CYCLOHEXANE-1,3-DIOL, ARG-HIS-LYS-ILE-URL-URK-URL-LEU-GLN, Vitamin D3 receptor A | | Authors: | Buratto, J, Belorusova, A.Y, Rochel, N, Guichard, G. | | Deposit date: | 2020-02-04 | | Release date: | 2021-02-17 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.372 Å) | | Cite: | Structural Basis for alpha-Helix Mimicry and Inhibition of Protein-Protein Interactions with Oligourea Foldamers.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
6XZI
 
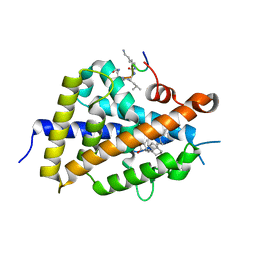 | | Structure of zVDR LBD-calcitriol in complex with chimera 11 | | Descriptor: | 5-{2-[1-(5-HYDROXY-1,5-DIMETHYL-HEXYL)-7A-METHYL-OCTAHYDRO-INDEN-4-YLIDENE]-ETHYLIDENE}-4-METHYLENE-CYCLOHEXANE-1,3-DIOL, ACETATE ION, ARG-HIS-LYS-ILE-LEU-URK-UIL-URL, ... | | Authors: | Buratto, J, Belorusova, A.Y, Rochel, N, Guichard, G. | | Deposit date: | 2020-02-04 | | Release date: | 2021-02-17 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Basis for alpha-Helix Mimicry and Inhibition of Protein-Protein Interactions with Oligourea Foldamers.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
6XZK
 
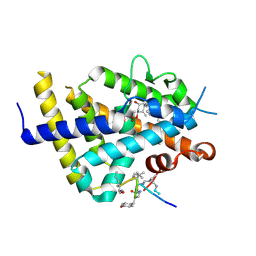 | | Structure of zVDR LBD-Calcitriol in complex with chimera 13 | | Descriptor: | 5-{2-[1-(5-HYDROXY-1,5-DIMETHYL-HEXYL)-7A-METHYL-OCTAHYDRO-INDEN-4-YLIDENE]-ETHYLIDENE}-4-METHYLENE-CYCLOHEXANE-1,3-DIOL, ACETATE ION, GLU-ASN-ALA-UIA-URL-URY-URV-UZN-LYS, ... | | Authors: | Buratto, J, Belorusova, A.Y, Rochel, N, Guichard, G. | | Deposit date: | 2020-02-04 | | Release date: | 2021-02-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Basis for alpha-Helix Mimicry and Inhibition of Protein-Protein Interactions with Oligourea Foldamers.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
6XZV
 
 | | Structure of zVDR LBD-Calcitriol in complex with chimera 18 | | Descriptor: | 5-{2-[1-(5-HYDROXY-1,5-DIMETHYL-HEXYL)-7A-METHYL-OCTAHYDRO-INDEN-4-YLIDENE]-ETHYLIDENE}-4-METHYLENE-CYCLOHEXANE-1,3-DIOL, URA-UIA-URL-URY-URV-UZN-LYS, Vitamin D3 receptor A | | Authors: | Buratto, J, Belorusova, A.Y, Rochel, N, Guichard, G. | | Deposit date: | 2020-02-05 | | Release date: | 2021-02-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Basis for alpha-Helix Mimicry and Inhibition of Protein-Protein Interactions with Oligourea Foldamers.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
1CWZ
 
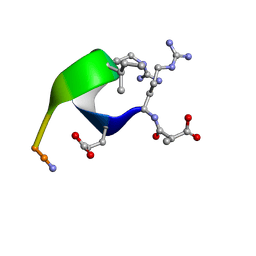 | | Solution structure of the analogue retro-inverso (MA-S)REGRIGGC in contact with the monoclonal antibody MAB 4X11, NMR, 7 structures | | Descriptor: | HISTONE H3, METHYLMALONIC ACID | | Authors: | Phan Chan Du, A, Petit, M.C, Guichard, G, Briand, J.P, Muller, S, Cung, M.T. | | Deposit date: | 1999-08-27 | | Release date: | 1999-09-03 | | Last modified: | 2024-10-09 | | Method: | SOLUTION NMR | | Cite: | Structure of antibody-bound peptides and retro-inverso analogues. A transferred nuclear Overhauser effect spectroscopy and molecular dynamics approach.
Biochemistry, 40, 2001
|
|
6ZUF
 
 | | Urea-based Foldamer Inhibitor chimera C2 in complex with ASF1 Histone chaperone | | Descriptor: | C2 foldamer/peptide hybrid inhibitor of histone chaperone ASF1, GLYCEROL, Histone chaperone ASF1A, ... | | Authors: | Bakail, M, Mbianda, J, Perrin, E.M, Guerois, R, Legrand, P, Traore, S, Douat, C, Guichard, G, Ochsenbein, F. | | Deposit date: | 2020-07-22 | | Release date: | 2021-06-09 | | Last modified: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (1.798 Å) | | Cite: | Optimal anchoring of a foldamer inhibitor of ASF1 histone chaperone through backbone plasticity.
Sci Adv, 7, 2021
|
|
1CW8
 
 | | SOLUTION STRUCTURE OF THE ANALOGUE RETRO-INVERSO (mA-R)REGRIGGC IN CONTACT WITH THE MONOCLONAL ANTIBODY MAB 4X11, NMR, 6 STRUCTURES | | Descriptor: | HISTONE H3, METHYLMALONIC ACID | | Authors: | Phan Chan Du, A, Petit, M.C, Guichard, G, Briand, J.P, Muller, S, Cung, T. | | Deposit date: | 1999-08-26 | | Release date: | 1999-09-03 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | Structure of antibody-bound peptides and retro-inverso analogues. A transferred nuclear Overhauser effect spectroscopy and molecular dynamics approach.
Biochemistry, 40, 2001
|
|
1CT6
 
 | | SOLUTION STRUCTURE OF CGGIRGERG IN CONTACT WITH THE MONOCLONAL ANTIBODY MAB 4X11, NMR, 11 STRUCTURES | | Descriptor: | HISTONE H3 PEPTIDE | | Authors: | Phan Chan Du, A, Petit, M.C, Guichard, G, Briand, J.P, Muller, S, Cung, M.T. | | Deposit date: | 1999-08-19 | | Release date: | 1999-09-02 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure of antibody-bound peptides and retro-inverso analogues. A transferred nuclear Overhauser effect spectroscopy and molecular dynamics approach.
Biochemistry, 40, 2001
|
|
1CS9
 
 | | SOLUTION STRUCTURE OF CGGIRGERA IN CONTACT WITH THE MONOCLONAL ANTIBODY MAB 4X11, NMR, 7 STRUCTURES | | Descriptor: | HISTONE H3 PEPTIDE | | Authors: | Phan Chan Du, A, Petit, M.C, Guichard, G, Briand, J.P, Muller, S, Cung, M.T. | | Deposit date: | 1999-08-18 | | Release date: | 1999-09-02 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure of antibody-bound peptides and retro-inverso analogues. A transferred nuclear Overhauser effect spectroscopy and molecular dynamics approach.
Biochemistry, 40, 2001
|
|
7AZ5
 
 | | DNA polymerase sliding clamp from Escherichia coli with peptide 47 bound | | Descriptor: | Beta sliding clamp, DI(HYDROXYETHYL)ETHER, Peptide 47, ... | | Authors: | Monsarrat, C, Compain, G, Andre, C, Martiel, I, Engilberge, S, Olieric, V, Wolff, P, Brillet, K, Landolfo, M, Silva da Veiga, C, Wagner, J, Guichard, G, Burnouf, D.Y. | | Deposit date: | 2020-11-16 | | Release date: | 2021-12-01 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Iterative Structure-Based Optimization of Short Peptides Targeting the Bacterial Sliding Clamp.
J.Med.Chem., 64, 2021
|
|
7AZG
 
 | | DNA polymerase sliding clamp from Escherichia coli with peptide 4 bound | | Descriptor: | Beta sliding clamp, Peptide 4 | | Authors: | Monsarrat, C, Compain, G, Andre, C, Martiel, I, Engilberge, S, Olieric, V, Wolff, P, Brillet, K, Landolfo, M, Silva da Veiga, C, Wagner, J, Guichard, G, Burnouf, D.Y. | | Deposit date: | 2020-11-16 | | Release date: | 2021-12-01 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.92 Å) | | Cite: | Iterative Structure-Based Optimization of Short Peptides Targeting the Bacterial Sliding Clamp.
J.Med.Chem., 64, 2021
|
|
