6GHW
 
 | | Substituting the prolines of 4-oxalocrotonate tautomerase with non-canonical analogue (2S)-3,4-dehydroproline | | Descriptor: | 2-hydroxymuconate tautomerase, CALCIUM ION | | Authors: | Pavkov-Keller, T, Lukesch, M.S, Wiltschi, B, Gruber, K. | | Deposit date: | 2018-05-09 | | Release date: | 2019-03-06 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Substituting the catalytic proline of 4-oxalocrotonate tautomerase with non-canonical analogues reveals a finely tuned catalytic system.
Sci Rep, 9, 2019
|
|
5E4B
 
 | |
5E46
 
 | |
3RLI
 
 | | Crystal structure of monoacylglycerol lipase from Bacillus sp. H257 in complex with PMSF | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, Thermostable monoacylglycerol lipase, phenylmethanesulfonic acid | | Authors: | Rengachari, S, Bezerra, G.A, Gruber, K, Oberer, M. | | Deposit date: | 2011-04-19 | | Release date: | 2012-05-23 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.854 Å) | | Cite: | The structure of monoacylglycerol lipase from Bacillus sp. H257 reveals unexpected conservation of the cap architecture between bacterial and human enzymes.
Biochim.Biophys.Acta, 1821, 2012
|
|
3RM3
 
 | | Crystal structure of monoacylglycerol lipase from Bacillus sp. H257 | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, Thermostable monoacylglycerol lipase | | Authors: | Rengachari, S, Bezerra, G.A, Gruber, K, Oberer, M. | | Deposit date: | 2011-04-20 | | Release date: | 2012-05-02 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | The structure of monoacylglycerol lipase from Bacillus sp. H257 reveals unexpected conservation of the cap architecture between bacterial and human enzymes.
Biochim.Biophys.Acta, 1821, 2012
|
|
4ZXF
 
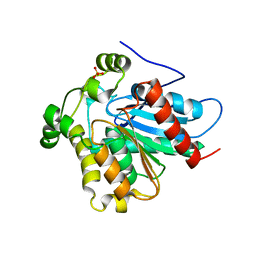 | | Crystal Structure of a Soluble Variant of Monoglyceride Lipase from Saccharomyces Cerevisiae in Complex with a Substrate Analog | | Descriptor: | 1-{3-[(R)-hydroxy(octadecyloxy)phosphoryl]propyl}triaza-1,2-dien-2-ium, Monoglyceride lipase, NITRATE ION, ... | | Authors: | Aschauer, P, Lichtenegger, J, Rengachari, S, Gruber, K, Oberer, M. | | Deposit date: | 2015-05-20 | | Release date: | 2016-05-25 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of the Saccharomyces cerevisiae monoglyceride lipase Yju3p.
Biochim.Biophys.Acta, 1861, 2016
|
|
3AX4
 
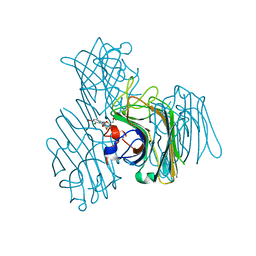 | | Three-dimensional structure of lectin from Dioclea violacea and comparative vasorelaxant effects with Dioclea rostrata | | Descriptor: | 5-bromo-4-chloro-1H-indol-3-yl alpha-D-mannopyranoside, CALCIUM ION, MANGANESE (II) ION, ... | | Authors: | Bezerra, M.J.B, Bezerra, G.A, Martins, J.L, Nascimento, K.S, Nagano, C.S, Gruber, K, Assereuy, A.M, Delatorre, P, Rocha, B.A.M, Cavada, B.S. | | Deposit date: | 2011-03-29 | | Release date: | 2012-04-04 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.613 Å) | | Cite: | Crystal structure of Dioclea violacea lectin and a comparative study of vasorelaxant properties with Dioclea rostrata lectin
Int.J.Biochem.Cell Biol., 45, 2013
|
|
4V3B
 
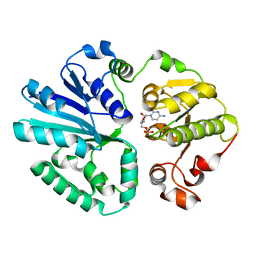 | | The structure of alpha2,3-sialyltransferase variant 1 from Pasteurella dagmatis in complex with the donor product CMP | | Descriptor: | CYTIDINE-5'-MONOPHOSPHATE, SIALYLTRANSFERASE | | Authors: | Pavkov-Keller, T, Schmoelzer, K, Czabany, T, Luley-Goedl, C, Ribitsch, D, Schwab, H, Nidetzky, B, Gruber, K. | | Deposit date: | 2014-10-17 | | Release date: | 2015-04-08 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Complete Switch from Alpha2,3- to Alpha2,6-Regioselectivity in Pasteurella Dagmatis Beta-D-Galactoside Sialyltransferase by Active-Site Redesign
Chem.Commun.(Camb.), 51, 2015
|
|
4V3C
 
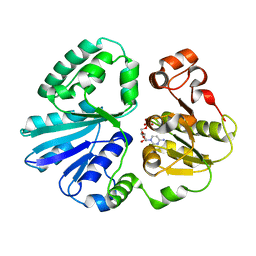 | | The structure of alpha2,3-sialyltransferase variant 2 from Pasteurella dagmatis in complex with the donor product CMP | | Descriptor: | CYTIDINE-5'-MONOPHOSPHATE, SIALYLTRANSFERASE | | Authors: | Pavkov-Keller, T, Schmoelzer, K, Czabany, T, Luley-Goedl, C, Ribitsch, D, Schwab, H, Nidetzky, B, Gruber, K. | | Deposit date: | 2014-10-17 | | Release date: | 2015-04-08 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Complete Switch from Alpha2,3- to Alpha2,6-Regioselectivity in Pasteurella Dagmatis Beta-D-Galactoside Sialyltransferase by Active-Site Redesign
Chem.Commun.(Camb.), 51, 2015
|
|
4V39
 
 | | Apo-structure of alpha2,3-sialyltransferase variant 2 from Pasteurella dagmatis | | Descriptor: | SIALYLTRANSFERASE | | Authors: | Pavkov-Keller, T, Schmoelzer, K, Czabany, T, Luley-Goedl, C, Ribitsch, D, Schwab, H, Nidetzky, B, Gruber, K. | | Deposit date: | 2014-10-17 | | Release date: | 2015-04-08 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Complete Switch from Alpha2,3- to Alpha2,6-Regioselectivity in Pasteurella Dagmatis Beta-D-Galactoside Sialyltransferase by Active-Site Redesign
Chem.Commun.(Camb.), 51, 2015
|
|
2XAA
 
 | | Alcohol dehydrogenase ADH-'A' from Rhodococcus ruber DSM 44541 at pH 8.5 in complex with NAD and butane-1,4-diol | | Descriptor: | 1,4-BUTANEDIOL, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, SECONDARY ALCOHOL DEHYDROGENASE, ... | | Authors: | Kroutil, W, Gruber, K, Grogan, G. | | Deposit date: | 2010-03-30 | | Release date: | 2010-08-11 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural Insights Into Substrate Specificity and Solvent Tolerance in Alcohol Dehydrogenase Adh-'A' from Rhodococcus Ruber Dsm 44541.
Chem.Commun.(Camb.), 46, 2010
|
|
4UD8
 
 | | AtBBE15 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 3,6,9,12,15,18,21,24-OCTAOXAHEXACOSAN-1-OL, ... | | Authors: | Daniel, B, Steiner, B, Pavkov-Keller, T, Dordic, A, Gutmann, A, Sensen, C.W, Nidetzky, B, van der Graaff, E, Wallner, S, Gruber, K, Macheroux, P. | | Deposit date: | 2014-12-09 | | Release date: | 2015-06-10 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.088 Å) | | Cite: | Oxidation of Monolignols by Members of the Berberine Bridge Enzyme Family Suggests a Role in Cell Wall Metabolism.
J.Biol.Chem., 290, 2015
|
|
4V38
 
 | | Apo-structure of alpha2,3-sialyltransferase variant 1 from Pasteurella dagmatis | | Descriptor: | SIALYLTRANSFERASE | | Authors: | Pavkov-Keller, T, Schmoelzer, K, Czabany, T, Luley-Goedl, C, Ribitsch, D, Schwab, H, Nidetzky, B, Gruber, K. | | Deposit date: | 2014-10-17 | | Release date: | 2015-04-08 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Complete Switch from Alpha2,3- to Alpha2,6-Regioselectivity in Pasteurella Dagmatis Beta-D-Galactoside Sialyltransferase by Active-Site Redesign
Chem.Commun.(Camb.), 51, 2015
|
|
1CCW
 
 | |
4WJL
 
 | | Structure of human dipeptidyl peptidase 10 (DPPY): a modulator of neuronal Kv4 channels | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Inactive dipeptidyl peptidase 10, ... | | Authors: | Bezerra, G.A, Dobrovetsky, E, Seitova, A, Fedosyuk, S, Dhe-Paganon, S, Gruber, K. | | Deposit date: | 2014-09-30 | | Release date: | 2015-03-18 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Structure of human dipeptidyl peptidase 10 (DPPY): a modulator of neuronal Kv4 channels.
Sci Rep, 5, 2015
|
|
4U2A
 
 | | Structure of a lectin from the seeds of Vatairea macrocarpa complexed with GalNAc | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-galactopyranose, CALCIUM ION, CITRIC ACID, ... | | Authors: | Sousa, B.L, Silva-Filho, J.C, Kumar, P, Lyskowski, A, Bezerra, G.A, Delatorre, P, Rocha, B.A.M, Nagano, C.S, Gruber, K, Cavada, B.S. | | Deposit date: | 2014-07-16 | | Release date: | 2014-12-31 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | High-resolution structure of a new Tn antigen-binding lectin from Vatairea macrocarpa and a comparative analysis of Tn-binding legume lectins.
Int.J.Biochem.Cell Biol., 59C, 2014
|
|
4U36
 
 | | Crystal structure of a seed lectin from Vatairea macrocarpa complexed with Tn-antigen | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-galactopyranose, CALCIUM ION, CITRIC ACID, ... | | Authors: | Sousa, B.L, Silva-Filho, J.C, Kumar, P, Lyskowski, A, Bezerra, G.A, Delatorre, P, Rocha, B.A.M, Nagano, C.S, Gruber, K, Cavada, B.S. | | Deposit date: | 2014-07-18 | | Release date: | 2014-12-31 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | High-resolution structure of a new Tn antigen-binding lectin from Vatairea macrocarpa and a comparative analysis of Tn-binding legume lectins.
Int.J.Biochem.Cell Biol., 59C, 2014
|
|
4CET
 
 | | Crystal structure of the complex of the P187S variant of human NAD(P) H:quinone oxidoreductase with dicoumarol at 2.2 A resolution | | Descriptor: | BISHYDROXY[2H-1-BENZOPYRAN-2-ONE,1,2-BENZOPYRONE], FLAVIN-ADENINE DINUCLEOTIDE, NAD(P)H DEHYDROGENASE [QUINONE] 1 | | Authors: | Lienhart, W.D, Gudipati, V, Uhl, M.K, Binter, A, Pulido, S, Saf, R, Zangger, K, Gruber, K, Macheroux, P. | | Deposit date: | 2013-11-12 | | Release date: | 2014-08-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Collapse of the Native Structure by a Single Amino Acid Exchange in Human Nad(P)H:Quinone Oxidoreductase (Nqo1).
FEBS J., 281, 2014
|
|
4CF6
 
 | | Crystal structure of the complex of the P187S variant of human NAD(P) H:quinone oxidoreductase with Cibacron blue at 2.7 A resolution | | Descriptor: | CIBACRON BLUE, FLAVIN-ADENINE DINUCLEOTIDE, NAD(P)H DEHYDROGENASE [QUINONE] 1 | | Authors: | Lienhart, W.D, Gudipati, V, Uhl, M.K, Binter, A, Pulido, S, Saf, R, Zangger, K, Gruber, K, Macheroux, P. | | Deposit date: | 2013-11-13 | | Release date: | 2014-08-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.694 Å) | | Cite: | Collapse of the Native Structure by a Single Amino Acid Exchange in Human Nad(P)H:Quinone Oxidoreductase (Nqo1).
FEBS J., 281, 2014
|
|
5K3W
 
 | | Structural characterisation of fold IV-transaminase, CpuTA1, from Curtobacterium pusillum | | Descriptor: | 3-AMINOBENZOIC ACID, CpuTA1, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Pavkov-Keller, T, Diepold, M, Steiner, K, Gruber, K. | | Deposit date: | 2016-05-20 | | Release date: | 2016-12-14 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.503 Å) | | Cite: | Discovery and structural characterisation of new fold type IV-transaminases exemplify the diversity of this enzyme fold.
Sci Rep, 6, 2016
|
|
6FRI
 
 | |
6GG2
 
 | | The structure of FsqB from Aspergillus fumigatus, a flavoenzyme of the amine oxidase family | | Descriptor: | Amino acid oxidase fmpA, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Pavkov-Keller, T, Lahham, M, Macheroux, P, Gruber, K. | | Deposit date: | 2018-05-02 | | Release date: | 2018-09-19 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.598 Å) | | Cite: | Oxidative cyclization ofN-methyl-dopa by a fungal flavoenzyme of the amine oxidase family.
J. Biol. Chem., 293, 2018
|
|
4UIR
 
 | | Structure of oleate hydratase from Elizabethkingia meningoseptica | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, HEXAETHYLENE GLYCOL, OLEATE HYDRATASE, ... | | Authors: | Pavkov-Keller, T, Hromic, A, Engleder, M, Emmerstorfer, A, Steinkellner, G, Schrempf, S, Wriessnegger, T, Leitner, E, Strohmeier, G.A, Kaluzna, I, Mink, D, Schuermann, M, Wallner, S, Macheroux, P, Pichler, H, Gruber, K. | | Deposit date: | 2015-04-02 | | Release date: | 2015-07-01 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structure-Based Mechanism of Oleate Hydratase from Elizabethkingia Meningoseptica.
Chembiochem, 16, 2015
|
|
4D1Y
 
 | | Crystal structure of a putative protease from Bacteroides thetaiotaomicron. | | Descriptor: | PUTATIVE PROTEASE I, RIBOFLAVIN, ZINC ION | | Authors: | Knaus, T, Uhl, M.K, Monschein, S, Moratti, S, Gruber, K, Macheroux, P. | | Deposit date: | 2014-05-05 | | Release date: | 2014-10-22 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure and Stability of an Unusual Zinc-Binding Protein from Bacteroides Thetaiotaomicron.
Biochim.Biophys.Acta, 1844, 2014
|
|
4D7K
 
 | |
