1SCI
 
 | | K236L mutant of hydroxynitrile lyase from Hevea brasiliensis | | Descriptor: | (S)-acetone-cyanohydrin lyase, SULFATE ION | | Authors: | Gruber, K, Gartler, G, Krammer, B, Schwab, H, Kratky, C. | | Deposit date: | 2004-02-12 | | Release date: | 2004-06-29 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Reaction mechanism of hydroxynitrile lyases of the alpha/beta-hydrolase superfamily: the three-dimensional structure of the transient enzyme-substrate complex certifies the crucial role of LYS236
J.Biol.Chem., 279, 2004
|
|
1SCK
 
 | | K236L mutant of hydroxynitrile lyase from Hevea brasiliensis in complex with acetone | | Descriptor: | (S)-acetone-cyanohydrin lyase, ACETONE, SULFATE ION | | Authors: | Gruber, K, Gartler, G, Krammer, B, Schwab, H, Kratky, C. | | Deposit date: | 2004-02-12 | | Release date: | 2004-06-29 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Reaction mechanism of hydroxynitrile lyases of the alpha/beta-hydrolase superfamily: the three-dimensional structure of the transient enzyme-substrate complex certifies the crucial role of LYS236
J.Biol.Chem., 279, 2004
|
|
1SCQ
 
 | | K236L mutant of hydroxynitrile lyase from Hevea brasiliensis in complex with acetonecyanohydrin | | Descriptor: | (S)-acetone-cyanohydrin lyase, 2-HYDROXY-2-METHYLPROPANENITRILE, SULFATE ION | | Authors: | Gruber, K, Gartler, G, Krammer, B, Schwab, H, Kratky, C. | | Deposit date: | 2004-02-12 | | Release date: | 2004-06-29 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Reaction mechanism of hydroxynitrile lyases of the alpha/beta-hydrolase superfamily: the three-dimensional structure of the transient enzyme-substrate complex certifies the crucial role of LYS236
J.Biol.Chem., 279, 2004
|
|
1SC9
 
 | | Hydroxynitrile Lyase from Hevea brasiliensis in complex with the natural substrate acetone cyanohydrin | | Descriptor: | (S)-acetone-cyanohydrin lyase, 2-HYDROXY-2-METHYLPROPANENITRILE, SULFATE ION | | Authors: | Gruber, K, Gartler, G, Krammer, B, Schwab, H, Kratky, C. | | Deposit date: | 2004-02-12 | | Release date: | 2004-06-29 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Reaction mechanism of hydroxynitrile lyases of the alpha/beta-hydrolase superfamily: the three-dimensional structure of the transient enzyme-substrate complex certifies the crucial role of LYS236
J.Biol.Chem., 279, 2004
|
|
1I9C
 
 | |
1YB6
 
 | |
1YB7
 
 | | Hydroxynitrile lyase from hevea brasiliensis in complex with 2,3-dimethyl-2-hydroxy-butyronitrile | | Descriptor: | (S)-2-HYDROXY-2,3-DIMETHYLBUTANENITRILE, (S)-acetone-cyanohydrin lyase, SULFATE ION | | Authors: | Gruber, K, Gartler, G, Kratky, C. | | Deposit date: | 2004-12-20 | | Release date: | 2005-12-20 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Structural determinants of the enantioselectivity of the hydroxynitrile lyase from Hevea brasiliensis
J.Biotechnol., 129, 2007
|
|
15C8
 
 | | CATALYTIC ANTIBODY 5C8, FREE FAB | | Descriptor: | IGG 5C8 FAB (HEAVY CHAIN), IGG 5C8 FAB (LIGHT CHAIN) | | Authors: | Gruber, K, Wilson, I.A. | | Deposit date: | 1998-03-18 | | Release date: | 1999-03-23 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Ligand-Induced Conformational Changes in a Catalytic Antibody: Comparison of the Bound and Unbound Structure of Fab 5C8
To be Published
|
|
35C8
 
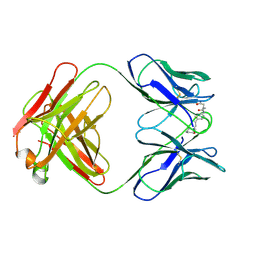 | | CATALYTIC ANTIBODY 5C8, FAB-INHIBITOR COMPLEX | | Descriptor: | IGG 5C8, N-(PARA-GLUTARAMIDOPHENYL-ETHYL)-PIPERIDINIUM-N-OXIDE | | Authors: | Gruber, K, Wilson, I.A. | | Deposit date: | 1998-03-18 | | Release date: | 1999-03-23 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for antibody catalysis of a disfavored ring closure reaction.
Biochemistry, 38, 1999
|
|
25C8
 
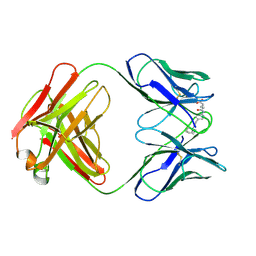 | | CATALYTIC ANTIBODY 5C8, FAB-HAPTEN COMPLEX | | Descriptor: | IGG 5C8, N-METHYL-N-(PARA-GLUTARAMIDOPHENYL-ETHYL)-PIPERIDINIUM ION | | Authors: | Gruber, K, Wilson, I.A. | | Deposit date: | 1998-03-18 | | Release date: | 1999-03-23 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for antibody catalysis of a disfavored ring closure reaction.
Biochemistry, 38, 1999
|
|
6FY4
 
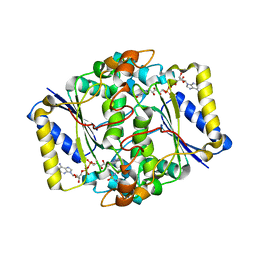 | |
1CB7
 
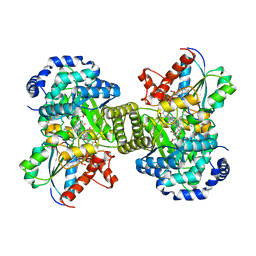 | |
6H9F
 
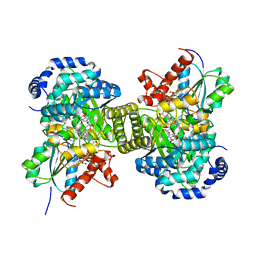 | | Structure of glutamate mutase reconstituted with bishomo-coenzyme B12 | | Descriptor: | (2~{R},3~{R},4~{S},5~{R})-2-(6-aminopurin-9-yl)-5-propyl-oxolane-3,4-diol, COBALAMIN, D(-)-TARTARIC ACID, ... | | Authors: | Gruber, K, Csitkovits, V, Kratky, C. | | Deposit date: | 2018-08-03 | | Release date: | 2019-08-14 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure-Based Demystification of Radical Catalysis by a Coenzyme B 12 Dependent Enzyme-Crystallographic Study of Glutamate Mutase with Cofactor Homologues.
Angew.Chem.Int.Ed.Engl., 61, 2022
|
|
6H9E
 
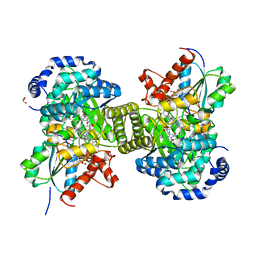 | | Structure of glutamate mutase reconstituted with homo-coenzyme B12 | | Descriptor: | (2~{R},3~{R},4~{S},5~{R})-2-(6-aminopurin-9-yl)-5-ethyl-oxolane-3,4-diol, COBALAMIN, D(-)-TARTARIC ACID, ... | | Authors: | Gruber, K, Csitkovits, V, Kratky, C. | | Deposit date: | 2018-08-03 | | Release date: | 2019-08-14 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Structure-Based Demystification of Radical Catalysis by a Coenzyme B 12 Dependent Enzyme-Crystallographic Study of Glutamate Mutase with Cofactor Homologues.
Angew.Chem.Int.Ed.Engl., 61, 2022
|
|
2YAS
 
 | |
1YNA
 
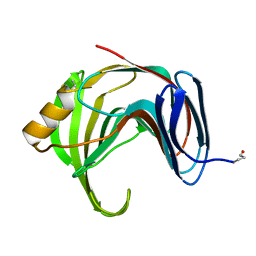 | | ENDO-1,4-BETA-XYLANASE, ROOM TEMPERATURE, PH 4.0 | | Descriptor: | ENDO-1,4-BETA-XYLANASE | | Authors: | Gruber, K, Kratky, C. | | Deposit date: | 1996-08-22 | | Release date: | 1997-02-12 | | Last modified: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Thermophilic xylanase from Thermomyces lanuginosus: high-resolution X-ray structure and modeling studies.
Biochemistry, 37, 1998
|
|
4UXA
 
 | | Improved variant of (R)-selective manganese-dependent hydroxynitrile lyase from bacteria | | Descriptor: | CUPIN 2 CONSERVED BARREL DOMAIN PROTEIN, MANGANESE (II) ION | | Authors: | Pavkov-Keller, T, Wiedner, R, Kothbauer, B, Gruber-Khadjawi, M, Schwab, H, Steiner, K, Gruber, K. | | Deposit date: | 2014-08-21 | | Release date: | 2015-01-21 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Improving the Properties of Bacterial R-Selective Hydroxynitrile Lyases for Industrial Applications
Chemcatchem, 2015
|
|
3ZOH
 
 | | Crystal structure of FMN-binding protein (YP_005476) from Thermus thermophilus with bound 1-Cyclohex-2-enone | | Descriptor: | FLAVIN MONONUCLEOTIDE, FLAVOREDOXIN, cyclohex-2-en-1-one | | Authors: | Pavkov-Keller, T, Steinkellner, G, Gruber, C.C, Steiner, K, Winkler, C, Schwamberger, O, Schwab, H, Faber, K, Gruber, K. | | Deposit date: | 2013-02-21 | | Release date: | 2014-05-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Identification of Promiscuous Ene-Reductase Activity by Mining Structural Databases Using Active Site Constellations.
Nat.Commun., 5, 2014
|
|
3ZOG
 
 | | Crystal structure of FMN-binding protein (NP_142786.1) from Pyrococcus horikoshii with bound 1-Cyclohex-2-enone | | Descriptor: | FLAVIN MONONUCLEOTIDE, FMN-BINDING PROTEIN, cyclohex-2-en-1-one | | Authors: | Pavkov-Keller, T, Steinkellner, G, Gruber, C.C, Steiner, K, Winkler, C, Schwamberger, O, Schwab, H, Faber, K, Gruber, K. | | Deposit date: | 2013-02-21 | | Release date: | 2014-05-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Identification of Promiscuous Ene-Reductase Activity by Mining Structural Databases Using Active Site Constellations.
Nat.Commun., 5, 2014
|
|
3ZOE
 
 | | Crystal structure of FMN-binding protein (YP_005476) from Thermus thermophilus with bound p-hydroxybenzaldehyde | | Descriptor: | FLAVIN MONONUCLEOTIDE, FLAVOREDOXIN, P-HYDROXYBENZALDEHYDE | | Authors: | Pavkov-Keller, T, Steinkellner, G, Gruber, C.C, Steiner, K, Winkler, C, Schwamberger, O, Schwab, H, Faber, K, Gruber, K. | | Deposit date: | 2013-02-21 | | Release date: | 2014-05-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Identification of Promiscuous Ene-Reductase Activity by Mining Structural Databases Using Active Site Constellations.
Nat.Commun., 5, 2014
|
|
3ZOD
 
 | | Crystal structure of FMN-binding protein (NP_142786.1) from Pyrococcus horikoshii with bound benzene-1,4-diol | | Descriptor: | FLAVIN MONONUCLEOTIDE, FMN-BINDING PROTEIN, benzene-1,4-diol | | Authors: | Pavkov-Keller, T, Steinkellner, G, Gruber, C.C, Steiner, K, Winkler, C, Schwamberger, O, Schwab, H, Faber, K, Gruber, K. | | Deposit date: | 2013-02-21 | | Release date: | 2014-05-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Identification of Promiscuous Ene-Reductase Activity by Mining Structural Databases Using Active Site Constellations.
Nat.Commun., 5, 2014
|
|
3ZOC
 
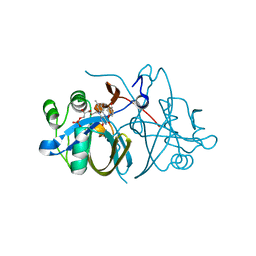 | | Crystal structure of FMN-binding protein (NP_142786.1) from Pyrococcus horikoshii with bound p-hydroxybenzaldehyde | | Descriptor: | FLAVIN MONONUCLEOTIDE, FMN-BINDING PROTEIN, P-HYDROXYBENZALDEHYDE | | Authors: | Pavkov-Keller, T, Steinkellner, G, Gruber, C.C, Steiner, K, Winkler, C, Schwamberger, O, Schwab, H, Faber, K, Gruber, K. | | Deposit date: | 2013-02-21 | | Release date: | 2014-05-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Identification of Promiscuous Ene-Reductase Activity by Mining Structural Databases Using Active Site Constellations.
Nat.Commun., 5, 2014
|
|
1QJ4
 
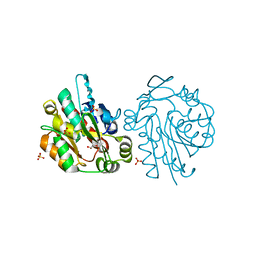 | |
4CE5
 
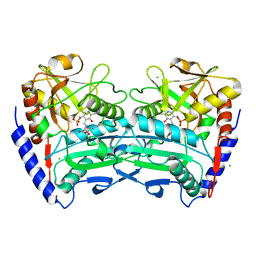 | | First crystal structure of an (R)-selective omega-transaminase from Aspergillus terreus | | Descriptor: | AT-OMEGATA, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Lyskowski, A, Gruber, C, Steinkellner, G, Schurmann, M, Schwab, H, Gruber, K, Steiner, K. | | Deposit date: | 2013-11-08 | | Release date: | 2014-02-12 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Crystal Structure of an (R)-Selective Omega-Transaminase from Aspergillus Terreus
Plos One, 9, 2014
|
|
3ZOF
 
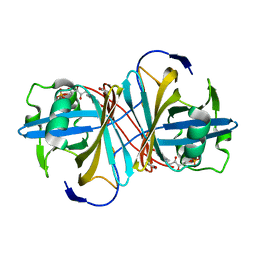 | | Crystal structure of FMN-binding protein (YP_005476) from Thermus thermophilus with bound benzene-1,4-diol | | Descriptor: | FLAVIN MONONUCLEOTIDE, FLAVOREDOXIN, benzene-1,4-diol | | Authors: | Pavkov-Keller, T, Steinkellner, G, Gruber, C.C, Steiner, K, Winkler, C, Schwamberger, O, Schwab, H, Faber, K, Gruber, K. | | Deposit date: | 2013-02-21 | | Release date: | 2014-05-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Identification of Promiscuous Ene-Reductase Activity by Mining Structural Databases Using Active Site Constellations.
Nat.Commun., 5, 2014
|
|
