8ADX
 
 | |
8B30
 
 | |
4KE6
 
 | | Crystal structure D196N mutant of Monoglyceride lipase from Bacillus sp. H257 in complex with 1-rac-lauroyl glycerol | | Descriptor: | (2R)-2,3-dihydroxypropyl dodecanoate, (4S)-2-METHYL-2,4-PENTANEDIOL, Thermostable monoacylglycerol lipase | | Authors: | Rengachari, S, Aschauer, P, Gruber, K, Dreveny, I, Oberer, M. | | Deposit date: | 2013-04-25 | | Release date: | 2013-09-18 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Conformational plasticity and ligand binding of bacterial monoacylglycerol lipase.
J.Biol.Chem., 288, 2013
|
|
3GDP
 
 | | Hydroxynitrile lyase from almond, monoclinic crystal form | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Dreveny, I, Gruber, K, Kratky, C. | | Deposit date: | 2009-02-24 | | Release date: | 2009-03-24 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Substrate binding in the FAD-dependent hydroxynitrile lyase from almond provides insight into the mechanism of cyanohydrin formation and explains the absence of dehydrogenation activity.
Biochemistry, 48, 2009
|
|
3T6J
 
 | |
3T6B
 
 | |
4K82
 
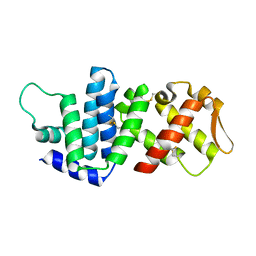 | | Crystal structure of lv-ranaspumin (Lv-RSN-1) from the foam nest of Leptodactylus vastus, monoclinic crystal form | | Descriptor: | Lv-ranaspumin (Lv-RSN-1) | | Authors: | Hissa, D.C, Bezerra, G.A, Melo, V.M.M, Gruber, K. | | Deposit date: | 2013-04-17 | | Release date: | 2014-03-05 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Unique Crystal Structure of a Novel Surfactant Protein from the Foam Nest of the Frog Leptodactylus vastus.
Chembiochem, 15, 2014
|
|
4KE7
 
 | | Crystal structure of Monoglyceride lipase from Bacillus sp. H257 in complex with an 1-myristoyl glycerol analogue | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Thermostable monoacylglycerol lipase, dodecyl hydrogen (S)-(3-azidopropyl)phosphonate | | Authors: | Rengachari, S, Aschauer, P, Gruber, K, Dreveny, I, Oberer, M. | | Deposit date: | 2013-04-25 | | Release date: | 2013-09-18 | | Last modified: | 2013-11-20 | | Method: | X-RAY DIFFRACTION (1.699 Å) | | Cite: | Conformational plasticity and ligand binding of bacterial monoacylglycerol lipase.
J.Biol.Chem., 288, 2013
|
|
3D2H
 
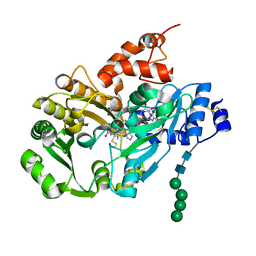 | | Structure of berberine bridge enzyme from Eschscholzia californica, monoclinic crystal form | | Descriptor: | (2R,3S,4S)-5-[(4R)-6',7'-dimethyl-2,3',5-trioxo-1'H-spiro[imidazolidine-4,2'-quinoxalin]-4'(3'H)-yl]-2,3,4-trihydroxypentyl-adenosine diphosphate, 2-acetamido-2-deoxy-beta-D-glucopyranose, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Winkler, A, Lyskowski, A, Macheroux, P, Gruber, K. | | Deposit date: | 2008-05-08 | | Release date: | 2008-10-28 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | A concerted mechanism for berberine bridge enzyme
Nat.Chem.Biol., 4, 2008
|
|
3D2D
 
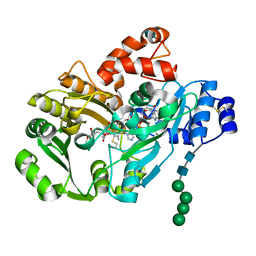 | | Structure of berberine bridge enzyme in complex with (S)-reticuline | | Descriptor: | (S)-reticuline, 2-acetamido-2-deoxy-beta-D-glucopyranose, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Winkler, A, Lyskowski, A, Macheroux, P, Gruber, K. | | Deposit date: | 2008-05-08 | | Release date: | 2008-10-28 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.796 Å) | | Cite: | A concerted mechanism for berberine bridge enzyme
Nat.Chem.Biol., 4, 2008
|
|
4K83
 
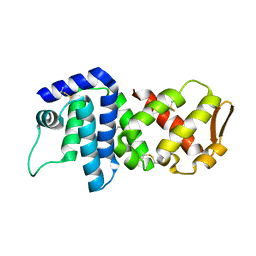 | | Crystal structure of lv-ranaspumin (Lv-RSN-1) from the foam nest of Leptodactylus vastus, orthorhombic crystal form | | Descriptor: | Lv-ranaspumin (Lv-RSN-1) | | Authors: | Hissa, D.C, Bezerra, G.A, Melo, V.M.M, Gruber, K. | | Deposit date: | 2013-04-17 | | Release date: | 2014-03-05 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Unique Crystal Structure of a Novel Surfactant Protein from the Foam Nest of the Frog Leptodactylus vastus.
Chembiochem, 15, 2014
|
|
2BK0
 
 | |
2WQL
 
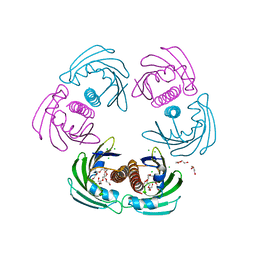 | | CRYSTAL STRUCTURE OF THE MAJOR CARROT ALLERGEN DAU C 1 | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, MAJOR ALLERGEN DAU C 1, ... | | Authors: | Markovic-Housley, Z, Basle, A, Padavattan, S, Hoffmann-Sommergruber, K, Schirmer, T. | | Deposit date: | 2009-08-24 | | Release date: | 2009-09-01 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure of the Major Carrot Allergen Dau C 1.
Acta Crystallogr.,Sect.D, 65, 2009
|
|
4XUW
 
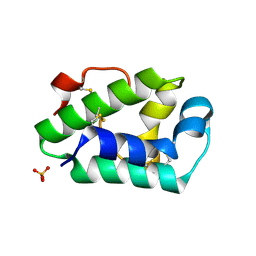 | | Structure of the hazelnut allergen, Cor a 8 | | Descriptor: | Non-specific lipid-transfer protein, PHOSPHATE ION | | Authors: | Offermann, L.R, Perdue, M.L, Bublin, M, Pfeifer, S, Dubiela, P, Hoffmann-Sommergruber, K, Chruszcz, M. | | Deposit date: | 2015-01-26 | | Release date: | 2015-10-14 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Structural and Functional Characterization of the Hazelnut Allergen Cor a 8.
J.Agric.Food Chem., 63, 2015
|
|
