6WFK
 
 | | Crystal structure of human Naa50 in complex with CoA and an inhibitor (compound 4a) identified using DNA encoded library technology | | Descriptor: | (4S)-1-methyl-N-{(3S,5S)-5-[4-(methylcarbamoyl)-1,3-thiazol-2-yl]-1-[4-(1H-tetrazol-5-yl)benzene-1-carbonyl]pyrrolidin-3-yl}-2,6-dioxohexahydropyrimidine-4-carboxamide, COENZYME A, N-alpha-acetyltransferase 50 | | Authors: | Greasley, S.E, Feng, J, Deng, Y.-L, Stewart, A.E. | | Deposit date: | 2020-04-03 | | Release date: | 2020-07-01 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Characterization of SpecificN-alpha-Acetyltransferase 50 (Naa50) Inhibitors Identified Using a DNA Encoded Library.
Acs Med.Chem.Lett., 11, 2020
|
|
1C3E
 
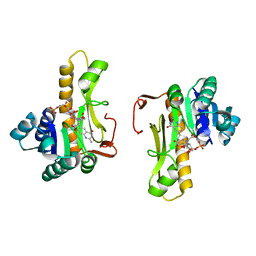 | | NEW INSIGHTS INTO INHIBITOR DESIGN FROM THE CRYSTAL STRUCTURE AND NMR STUDIES OF E. COLI GAR TRANSFORMYLATE IN COMPLEX WITH BETA-GAR AND 10-FORMYL-5,8,10-TRIDEAZAFOLIC ACID. | | Descriptor: | 2-{4-[2-(2-AMINO-4-HYDROXY-QUINAZOLIN-6-YL)-1-CARBOXY-ETHYL]-BENZOYLAMINO}-PENTANEDIOIC ACID, GLYCINAMIDE RIBONUCLEOTIDE, GLYCINAMIDE RIBONUCLEOTIDE TRANSFORMYLASE | | Authors: | Greasley, S.E, Yamashita, M.M, Cai, H, Benkovic, S.J, Boger, D.L, Wilson, I.A. | | Deposit date: | 1999-07-27 | | Release date: | 1999-12-29 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | New insights into inhibitor design from the crystal structure and NMR studies of Escherichia coli GAR transformylase in complex with beta-GAR and 10-formyl-5,8,10-trideazafolic acid.
Biochemistry, 38, 1999
|
|
1C2T
 
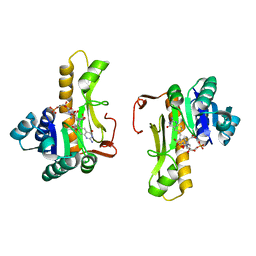 | | NEW INSIGHTS INTO INHIBITOR DESIGN FROM THE CRYSTAL STRUCTURE AND NMR STUDIES OF E. COLI GAR TRANSFORMYLASE IN COMPLEX WITH BETA-GAR AND 10-FORMYL-5,8,10-TRIDEAZAFOLIC ACID. | | Descriptor: | 10-FORMYL-5,8,10-TRIDEAZAFOLIC ACID, GLYCINAMIDE RIBONUCLEOTIDE, GLYCINAMIDE RIBONUCLEOTIDE TRANSFORMYLASE | | Authors: | Greasley, S.E, Yamashita, M.M, Cai, H, Benkovic, S.J, Boger, D.L, Wilson, I.A. | | Deposit date: | 1999-07-26 | | Release date: | 2000-01-05 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | New insights into inhibitor design from the crystal structure and NMR studies of Escherichia coli GAR transformylase in complex with beta-GAR and 10-formyl-5,8,10-trideazafolic acid.
Biochemistry, 38, 1999
|
|
1G8M
 
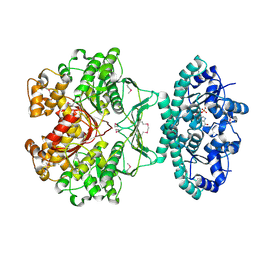 | | CRYSTAL STRUCTURE OF AVIAN ATIC, A BIFUNCTIONAL TRANSFORMYLASE AND CYCLOHYDROLASE ENZYME IN PURINE BIOSYNTHESIS AT 1.75 ANG. RESOLUTION | | Descriptor: | AICAR TRANSFORMYLASE-IMP CYCLOHYDROLASE, GUANOSINE-5'-MONOPHOSPHATE, POTASSIUM ION | | Authors: | Greasley, S.E, Horton, P, Beardsley, G.P, Benkovic, S.J, Wilson, I.A. | | Deposit date: | 2000-11-17 | | Release date: | 2001-04-27 | | Last modified: | 2017-10-04 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structure of a bifunctional transformylase and cyclohydrolase enzyme in purine biosynthesis.
Nat.Struct.Biol., 8, 2001
|
|
1RRG
 
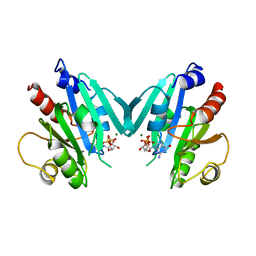 | | NON-MYRISTOYLATED RAT ADP-RIBOSYLATION FACTOR-1 COMPLEXED WITH GDP, DIMERIC CRYSTAL FORM | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, RAT ADP-RIBOSYLATION FACTOR-1 | | Authors: | Greasley, S.E, Jhoti, H, Bax, B. | | Deposit date: | 1995-12-16 | | Release date: | 1996-06-20 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The structure of rat ADP-ribosylation factor-1 (ARF-1) complexed to GDP determined from two different crystal forms.
Nat.Struct.Biol., 2, 1995
|
|
1RRF
 
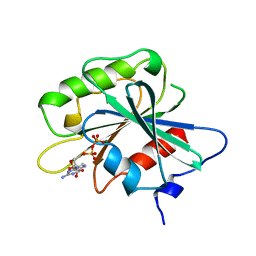 | | NON-MYRISTOYLATED RAT ADP-RIBOSYLATION FACTOR-1 COMPLEXED WITH GDP, MONOMERIC CRYSTAL FORM | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, RAT ADP-RIBOSYLATION FACTOR-1 | | Authors: | Greasley, S.E, Jhoti, H, Bax, B. | | Deposit date: | 1995-12-16 | | Release date: | 1996-06-20 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The structure of rat ADP-ribosylation factor-1 (ARF-1) complexed to GDP determined from two different crystal forms.
Nat.Struct.Biol., 2, 1995
|
|
3JYJ
 
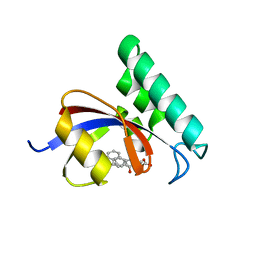 | | Structure-Based Design of Novel PIN1 Inhibitors (II) | | Descriptor: | (2R,4E)-2-[(naphthalen-2-ylcarbonyl)amino]-5-phenylpent-4-enoic acid, Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1 | | Authors: | Greasley, S.E, Ferre, R.A. | | Deposit date: | 2009-09-21 | | Release date: | 2010-04-07 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Structure-based design of novel human Pin1 inhibitors (II).
Bioorg.Med.Chem.Lett., 20, 2010
|
|
3I6C
 
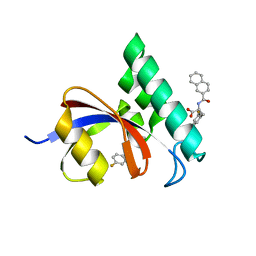 | | Structure-Based Design of Novel PIN1 Inhibitors (II) | | Descriptor: | 3-fluoro-N-(naphthalen-2-ylcarbonyl)-D-phenylalanine, Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1 | | Authors: | Greasley, S.E, Ferre, R.A. | | Deposit date: | 2009-07-06 | | Release date: | 2010-04-21 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structure-based design of novel human Pin1 inhibitors (II).
Bioorg.Med.Chem.Lett., 20, 2010
|
|
1JKX
 
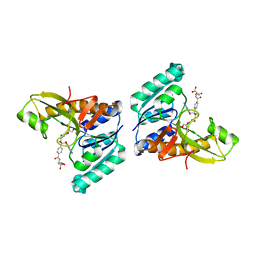 | | Unexpected formation of an epoxide-derived multisubstrate adduct inhibitor on the active site of GAR transformylase | | Descriptor: | N-[5'-O-PHOSPHONO-RIBOFURANOSYL]-2-[2-HYDROXY-2-[4-[GLUTAMIC ACID]-N-CARBONYLPHENYL]-3-[2-AMINO-4-HYDROXY-QUINAZOLIN-6-YL]-PROPANYLAMINO]-ACETAMIDE, PHOSPHORIBOSYLGLYCINAMIDE FORMYLTRANSFERASE | | Authors: | Greasley, S.E, Marsilje, T.H, Cai, H, Baker, S, Benkovic, S.J, Boger, D.L, Wilson, I.A. | | Deposit date: | 2001-07-13 | | Release date: | 2001-11-30 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Unexpected formation of an epoxide-derived multisubstrate adduct inhibitor on the active site of GAR transformylase.
Biochemistry, 40, 2001
|
|
4JPG
 
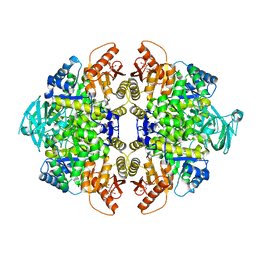 | | 2-((1H-benzo[d]imidazol-1-yl)methyl)-4H-pyrido[1,2-a]pyrimidin-4-ones as Novel PKM2 Activators | | Descriptor: | 1,6-di-O-phosphono-beta-D-fructofuranose, 2-(1H-benzimidazol-1-ylmethyl)-4H-pyrido[1,2-a]pyrimidin-4-one, Pyruvate kinase isozymes M1/M2 | | Authors: | Greasley, S.E, Hickey, M, Phonephaly, H, Cronin, C. | | Deposit date: | 2013-03-19 | | Release date: | 2013-05-22 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Discovery of 2-((1H-benzo[d]imidazol-1-yl)methyl)-4H-pyrido[1,2-a]pyrimidin-4-ones as novel PKM2 activators.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
3F7B
 
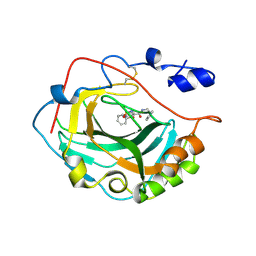 | | Crystal Structure of soluble domain of CA4 in complex with small molecule. | | Descriptor: | Carbonic anhydrase 4, N-(2-phenylethyl)-2-(phenylsulfanyl)-5-sulfamoylpyridine-3-carboxamide, ZINC ION | | Authors: | Greasley, S.E, Ferre, R.A.A, Pauly, T.A, Paz, R. | | Deposit date: | 2008-11-07 | | Release date: | 2009-09-22 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Thioether benzenesulfonamide inhibitors of carbonic anhydrases II and IV: structure-based drug design, synthesis, and biological evaluation.
Bioorg.Med.Chem., 18, 2010
|
|
3FW3
 
 | | Crystal Structure of soluble domain of CA4 in complex with Dorzolamide | | Descriptor: | (4S-TRANS)-4-(ETHYLAMINO)-5,6-DIHYDRO-6-METHYL-4H-THIENO(2,3-B)THIOPYRAN-2-SULFONAMIDE-7,7-DIOXIDE, Carbonic anhydrase 4, SULFATE ION, ... | | Authors: | Greasley, S.E, Ferre, R.A.A, Paz, R, Wickersham, J. | | Deposit date: | 2009-01-16 | | Release date: | 2009-12-01 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Thioether benzenesulfonamide inhibitors of carbonic anhydrases II and IV: structure-based drug design, synthesis, and biological evaluation.
Bioorg.Med.Chem., 18, 2010
|
|
3RWP
 
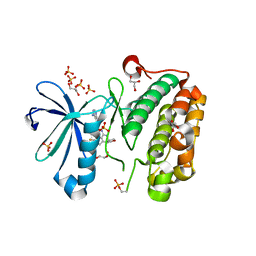 | | Discovery of a Novel, Potent and Selective Inhibitor of 3-Phosphoinositide Dependent Kinase (PDK1) | | Descriptor: | 3-phosphoinositide-dependent protein kinase 1, GLYCEROL, SULFATE ION, ... | | Authors: | Greasley, S.E, Hickey, M, Ferre, R.-A, Krauss, M, Cronin, C. | | Deposit date: | 2011-05-09 | | Release date: | 2011-11-16 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Discovery of Novel, Potent, and Selective Inhibitors of 3-Phosphoinositide-Dependent Kinase (PDK1).
J.Med.Chem., 54, 2011
|
|
3SC1
 
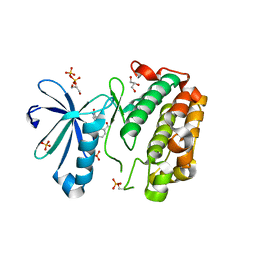 | | Novel Isoquinolone PDK1 Inhibitors Discovered through Fragment-Based Lead Discovery | | Descriptor: | 3-phosphoinositide-dependent protein kinase 1, 6-[2-(hydroxymethyl)phenyl]isoquinolin-1(2H)-one, GLYCEROL, ... | | Authors: | Greasley, S.E, Ferre, R.-A, Krauss, M, Cronin, C. | | Deposit date: | 2011-06-06 | | Release date: | 2011-10-26 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Novel isoquinolone PDK1 inhibitors discovered through fragment-based lead discovery.
J Comput Aided Mol Des, 25, 2011
|
|
1MEJ
 
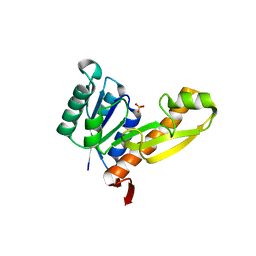 | | Human Glycinamide Ribonucleotide Transformylase domain at pH 8.5 | | Descriptor: | GLYCEROL, PHOSPHATE ION, Phosphoribosylglycinamide formyltransferase | | Authors: | Zhang, Y, Desharnais, J, Greasley, S.E, Beardsley, G.P, Boger, D.L, Wilson, I.A. | | Deposit date: | 2002-08-08 | | Release date: | 2002-12-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures of human GAR Tfase of low and high pH and with substrate beta-GAR
Biochemistry, 41, 2002
|
|
9AUL
 
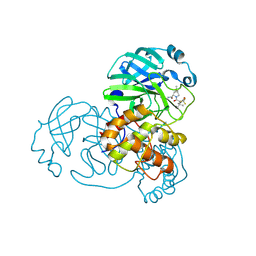 | | Structure of SARS-CoV-2 Mpro mutant (A173V,T304I)) in complex with Nirmatrelvir (PF-07321332) | | Descriptor: | (1R,2S,5S)-N-{(1E,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase nsp5 | | Authors: | Gajiwala, K.S, Greasley, S.E, Ferre, R.A, Liu, W, Stewart, A.E. | | Deposit date: | 2024-02-29 | | Release date: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (2.421 Å) | | Cite: | In vitro selection and analysis of SARS-CoV-2 nirmatrelvir resistance mutations contributing to clinical virus resistance surveillance.
Sci Adv, 10, 2024
|
|
9AUJ
 
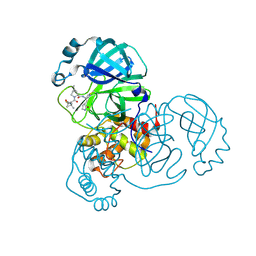 | | Structure of SARS-CoV-2 Mpro mutant (S144A) in complex with Nirmatrelvir (PF-07321332) | | Descriptor: | (1R,2S,5S)-N-{(1E,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase nsp5 | | Authors: | Gajiwala, K.S, Greasley, S.E, Ferre, R.A, Liu, W, Stewart, A.E. | | Deposit date: | 2024-02-29 | | Release date: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (1.486 Å) | | Cite: | In vitro selection and analysis of SARS-CoV-2 nirmatrelvir resistance mutations contributing to clinical virus resistance surveillance.
Sci Adv, 10, 2024
|
|
9AUM
 
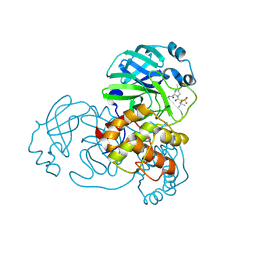 | | Structure of SARS-CoV-2 Mpro mutant (T21I,S144A,T304I) in complex with Nirmatrelvir (PF-07321332) | | Descriptor: | (1R,2S,5S)-N-{(1E,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase nsp5 | | Authors: | Gajiwala, K.S, Greasley, S.E, Ferre, R.A, Liu, W, Stewart, A.E. | | Deposit date: | 2024-02-29 | | Release date: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (1.539 Å) | | Cite: | In vitro selection and analysis of SARS-CoV-2 nirmatrelvir resistance mutations contributing to clinical virus resistance surveillance.
Sci Adv, 10, 2024
|
|
9AUK
 
 | | Structure of SARS-CoV-2 Mpro mutant (A173V) in complex with Nirmatrelvir (PF-07321332) | | Descriptor: | (1R,2S,5S)-N-{(1E,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase nsp5 | | Authors: | Gajiwala, K.S, Greasley, S.E, Ferre, R.A, Liu, W, Stewart, A.E. | | Deposit date: | 2024-02-29 | | Release date: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | In vitro selection and analysis of SARS-CoV-2 nirmatrelvir resistance mutations contributing to clinical virus resistance surveillance.
Sci Adv, 10, 2024
|
|
9AUO
 
 | | Structure of SARS-CoV-2 Mpro mutant (L50F,T304I) | | Descriptor: | 3C-like proteinase nsp5 | | Authors: | Gajiwala, K.S, Greasley, S.E, Ferre, R.A, Liu, W, Stewart, A.E. | | Deposit date: | 2024-02-29 | | Release date: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (2.423 Å) | | Cite: | In vitro selection and analysis of SARS-CoV-2 nirmatrelvir resistance mutations contributing to clinical virus resistance surveillance.
Sci Adv, 10, 2024
|
|
9AUN
 
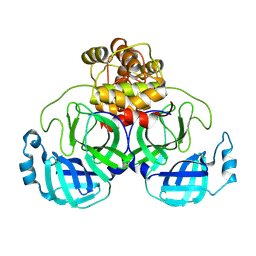 | | Structure of SARS-CoV-2 Mpro mutant (T21I,T304I) | | Descriptor: | 3C-like proteinase nsp5 | | Authors: | Gajiwala, K.S, Greasley, S.E, Ferre, R.A, Liu, W, Stewart, A.E. | | Deposit date: | 2024-02-29 | | Release date: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | In vitro selection and analysis of SARS-CoV-2 nirmatrelvir resistance mutations contributing to clinical virus resistance surveillance.
Sci Adv, 10, 2024
|
|
3OAW
 
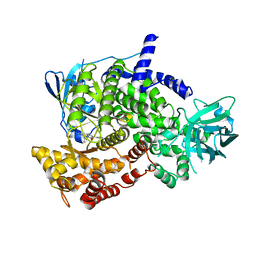 | |
4FA6
 
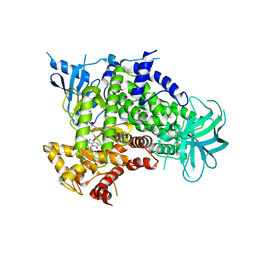 | |
2GAR
 
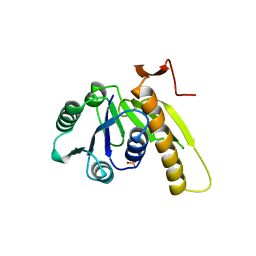 | | A PH-DEPENDENT STABLIZATION OF AN ACTIVE SITE LOOP OBSERVED FROM LOW AND HIGH PH CRYSTAL STRUCTURES OF MUTANT MONOMERIC GLYCINAMIDE RIBONUCLEOTIDE TRANSFORMYLASE | | Descriptor: | GLYCINAMIDE RIBONUCLEOTIDE TRANSFORMYLASE, PHOSPHATE ION | | Authors: | Su, Y, Yamashita, M.M, Greasley, S.E, Mullen, C.A, Shim, J.H, Jennings, P.A, Benkovic, S.J, Wilson, I.A. | | Deposit date: | 1998-05-13 | | Release date: | 1998-08-12 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A pH-dependent stabilization of an active site loop observed from low and high pH crystal structures of mutant monomeric glycinamide ribonucleotide transformylase at 1.8 to 1.9 A.
J.Mol.Biol., 281, 1998
|
|
3GAR
 
 | | A PH-DEPENDENT STABLIZATION OF AN ACTIVE SITE LOOP OBSERVED FROM LOW AND HIGH PH CRYSTAL STRUCTURES OF MUTANT MONOMERIC GLYCINAMIDE RIBONUCLEOTIDE TRANSFORMYLASE | | Descriptor: | GLYCINAMIDE RIBONUCLEOTIDE TRANSFORMYLASE, PHOSPHATE ION | | Authors: | Su, Y, Yamashita, M.M, Greasley, S.E, Mullen, C.A, Shim, J.H, Jennings, P.A, Benkovic, S.J, Wilson, I.A. | | Deposit date: | 1998-05-13 | | Release date: | 1998-08-12 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A pH-dependent stabilization of an active site loop observed from low and high pH crystal structures of mutant monomeric glycinamide ribonucleotide transformylase at 1.8 to 1.9 A.
J.Mol.Biol., 281, 1998
|
|
