1PKX
 
 | | Crystal Structure of human ATIC in complex with XMP | | Descriptor: | Bifunctional purine biosynthesis protein PURH, POTASSIUM ION, XANTHOSINE-5'-MONOPHOSPHATE | | Authors: | Wolan, D.W, Cheong, C.G, Greasley, S.E, Wilson, I.A. | | Deposit date: | 2003-06-06 | | Release date: | 2004-02-17 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Insights into the Human and Avian IMP Cyclohydrolase Mechanism via Crystal Structures with the Bound XMP Inhibitor.
Biochemistry, 43, 2004
|
|
3IKD
 
 | | Structure-Based Design of Novel PIN1 Inhibitors (I) | | Descriptor: | (2R)-2-[(1-benzothiophen-2-ylcarbonyl)amino]-3-phenylpropyl phosphate, Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1 | | Authors: | Matthews, D, Greasley, S, Ferre, R, Parge, H. | | Deposit date: | 2009-08-05 | | Release date: | 2009-09-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure-based design of novel human Pin1 inhibitors (I).
Bioorg.Med.Chem.Lett., 19, 2009
|
|
3GAR
 
 | | A PH-DEPENDENT STABLIZATION OF AN ACTIVE SITE LOOP OBSERVED FROM LOW AND HIGH PH CRYSTAL STRUCTURES OF MUTANT MONOMERIC GLYCINAMIDE RIBONUCLEOTIDE TRANSFORMYLASE | | Descriptor: | GLYCINAMIDE RIBONUCLEOTIDE TRANSFORMYLASE, PHOSPHATE ION | | Authors: | Su, Y, Yamashita, M.M, Greasley, S.E, Mullen, C.A, Shim, J.H, Jennings, P.A, Benkovic, S.J, Wilson, I.A. | | Deposit date: | 1998-05-13 | | Release date: | 1998-08-12 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A pH-dependent stabilization of an active site loop observed from low and high pH crystal structures of mutant monomeric glycinamide ribonucleotide transformylase at 1.8 to 1.9 A.
J.Mol.Biol., 281, 1998
|
|
1PL0
 
 | | Crystal structure of human ATIC in complex with folate-based inhibitor, BW2315U89UC | | Descriptor: | AMINOIMIDAZOLE 4-CARBOXAMIDE RIBONUCLEOTIDE, Bifunctional purine biosynthesis protein PURH, N-(4-{[(2-AMINO-4-OXO-3,4-DIHYDROQUINAZOLIN-6-YL)AMINO]SULFONYL}BENZOYL)GLUTAMIC ACID, ... | | Authors: | Cheong, C.G, Greasley, S.E, Horton, P.A, Beardsley, G.P, Wilson, I.A. | | Deposit date: | 2003-06-06 | | Release date: | 2004-04-27 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structures of Human Bifunctional Enzyme Aminoimidazole-4-carboxamide Ribonucleotide Transformylase/IMP Cyclohydrolase in Complex with Potent Sulfonyl-containing Antifolates.
J.Biol.Chem., 279, 2004
|
|
9AUN
 
 | | Structure of SARS-CoV-2 Mpro mutant (T21I,T304I) | | Descriptor: | 3C-like proteinase nsp5 | | Authors: | Gajiwala, K.S, Greasley, S.E, Ferre, R.A, Liu, W, Stewart, A.E. | | Deposit date: | 2024-02-29 | | Release date: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | In vitro selection and analysis of SARS-CoV-2 nirmatrelvir resistance mutations contributing to clinical virus resistance surveillance.
Sci Adv, 10, 2024
|
|
9AUK
 
 | | Structure of SARS-CoV-2 Mpro mutant (A173V) in complex with Nirmatrelvir (PF-07321332) | | Descriptor: | (1R,2S,5S)-N-{(1E,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase nsp5 | | Authors: | Gajiwala, K.S, Greasley, S.E, Ferre, R.A, Liu, W, Stewart, A.E. | | Deposit date: | 2024-02-29 | | Release date: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | In vitro selection and analysis of SARS-CoV-2 nirmatrelvir resistance mutations contributing to clinical virus resistance surveillance.
Sci Adv, 10, 2024
|
|
9AUO
 
 | | Structure of SARS-CoV-2 Mpro mutant (L50F,T304I) | | Descriptor: | 3C-like proteinase nsp5 | | Authors: | Gajiwala, K.S, Greasley, S.E, Ferre, R.A, Liu, W, Stewart, A.E. | | Deposit date: | 2024-02-29 | | Release date: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (2.423 Å) | | Cite: | In vitro selection and analysis of SARS-CoV-2 nirmatrelvir resistance mutations contributing to clinical virus resistance surveillance.
Sci Adv, 10, 2024
|
|
9AUL
 
 | | Structure of SARS-CoV-2 Mpro mutant (A173V,T304I)) in complex with Nirmatrelvir (PF-07321332) | | Descriptor: | (1R,2S,5S)-N-{(1E,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase nsp5 | | Authors: | Gajiwala, K.S, Greasley, S.E, Ferre, R.A, Liu, W, Stewart, A.E. | | Deposit date: | 2024-02-29 | | Release date: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (2.421 Å) | | Cite: | In vitro selection and analysis of SARS-CoV-2 nirmatrelvir resistance mutations contributing to clinical virus resistance surveillance.
Sci Adv, 10, 2024
|
|
9AUJ
 
 | | Structure of SARS-CoV-2 Mpro mutant (S144A) in complex with Nirmatrelvir (PF-07321332) | | Descriptor: | (1R,2S,5S)-N-{(1E,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase nsp5 | | Authors: | Gajiwala, K.S, Greasley, S.E, Ferre, R.A, Liu, W, Stewart, A.E. | | Deposit date: | 2024-02-29 | | Release date: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (1.486 Å) | | Cite: | In vitro selection and analysis of SARS-CoV-2 nirmatrelvir resistance mutations contributing to clinical virus resistance surveillance.
Sci Adv, 10, 2024
|
|
9AUM
 
 | | Structure of SARS-CoV-2 Mpro mutant (T21I,S144A,T304I) in complex with Nirmatrelvir (PF-07321332) | | Descriptor: | (1R,2S,5S)-N-{(1E,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase nsp5 | | Authors: | Gajiwala, K.S, Greasley, S.E, Ferre, R.A, Liu, W, Stewart, A.E. | | Deposit date: | 2024-02-29 | | Release date: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (1.539 Å) | | Cite: | In vitro selection and analysis of SARS-CoV-2 nirmatrelvir resistance mutations contributing to clinical virus resistance surveillance.
Sci Adv, 10, 2024
|
|
3IKG
 
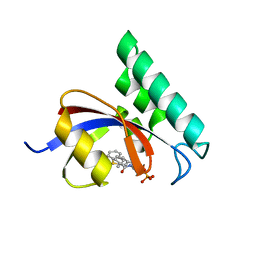 | | Structure-Based Design of Novel PIN1 Inhibitors (I) | | Descriptor: | (2R)-2-[(1-benzothiophen-2-ylcarbonyl)amino]-3-(3-methylphenyl)propyl phosphate, Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1 | | Authors: | Parge, H, Ferre, R.A, Greasley, S, Matthews, D. | | Deposit date: | 2009-08-05 | | Release date: | 2009-09-22 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Structure-based design of novel human Pin1 inhibitors (I).
Bioorg.Med.Chem.Lett., 19, 2009
|
|
1P4R
 
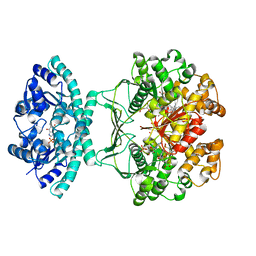 | | Crystal Structure of Human ATIC in complex with folate-based inhibitor BW1540U88UD | | Descriptor: | AMINOIMIDAZOLE 4-CARBOXAMIDE RIBONUCLEOTIDE, Bifunctional purine biosynthesis protein PURH, N-[(S)-(4-{[(2-AMINO-4-HYDROXYQUINAZOLIN-6-YL)(DIHYDROXY)-LAMBDA~4~-SULFANYL]AMINO}PHENYL)(HYDROXY)METHYL]-L-GLUTAMIC ACID, ... | | Authors: | Cheong, C.-G, Greasley, S.E, Horton, P.A, Beardsley, G.P, Wilson, I.A. | | Deposit date: | 2003-04-23 | | Release date: | 2004-03-30 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Crystal structures of human bifunctional enzyme aminoimidazole-4-carboxamide ribonucleotide transformylase/IMP cyclohydrolase in complex with potent sulfonyl-containing antifolates
J.Biol.Chem., 279, 2004
|
|
1OZ0
 
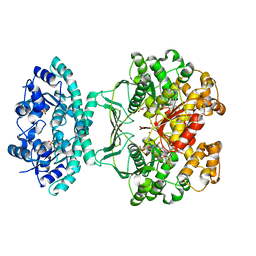 | | CRYSTAL STRUCTURE OF THE HOMODIMERIC BIFUNCTIONAL TRANSFORMYLASE AND CYCLOHYDROLASE ENZYME AVIAN ATIC IN COMPLEX WITH A MULTISUBSTRATE ADDUCT INHIBITOR BETA-DADF. | | Descriptor: | 2-[4-((2-AMINO-4-OXO-3,4-DIHYDRO-PYRIDO[3,2-D]PYRIMIDIN-6-YLMETHYL)-{3-[5-CARBAMOYL-3-(3,4- DIHYDROXY-5-PHOSPHONOOXYMETHYL-TETRAHYDRO-FURAN-2-YL)-3H-IMIDAZOL-4-YL]-ACRYLOYL}-AMINO)-BENZOYLAMINO]- PENTANEDIOIC ACID, Bifunctional purine biosynthesis protein PURH, PHOSPHATE ION, ... | | Authors: | Wolan, D.W, Greasley, S.E, Wall, M.J, Benkovic, S.J, Wilson, I.A. | | Deposit date: | 2003-04-07 | | Release date: | 2003-09-30 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of Avian AICAR Transformylase with a Multisubstrate Adduct Inhibitor beta-DADF Identifies the Folate Binding Site.
Biochemistry, 42, 2003
|
|
1D5T
 
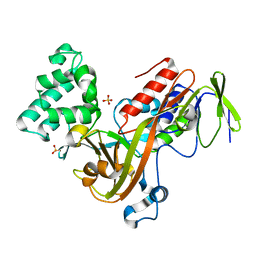 | | GUANINE NUCLEOTIDE DISSOCIATION INHIBITOR, ALPHA-ISOFORM | | Descriptor: | GUANINE NUCLEOTIDE DISSOCIATION INHIBITOR, SULFATE ION | | Authors: | Peng, L, Zeng, K, Heine, A, Moyer, B, Greasley, S.E, Kuhn, P, Balch, W.E, Wilson, I.A. | | Deposit date: | 1999-10-11 | | Release date: | 2000-10-25 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.04 Å) | | Cite: | A new functional domain of guanine nucleotide dissociation inhibitor (alpha-GDI) involved in Rab recycling.
Traffic, 1, 2000
|
|
