4D8I
 
 | |
4D8E
 
 | |
5O5O
 
 | | X-ray crystal structure of RapZ from Escherichia coli (P32 space group) | | Descriptor: | RNase adapter protein RapZ, SULFATE ION | | Authors: | Gonzalez, G.M, Durica-Mitic, S, Hardwick, S.W, Moncrieffe, M, Resch, M, Neumann, P, Ficner, R, Gorke, B, Luisi, B.F. | | Deposit date: | 2017-06-02 | | Release date: | 2017-08-30 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (3.404 Å) | | Cite: | Structural insights into RapZ-mediated regulation of bacterial amino-sugar metabolism.
Nucleic Acids Res., 45, 2017
|
|
5O5Q
 
 | | X-ray crystal structure of RapZ from Escherichia coli (P3221 space group) | | Descriptor: | RNase adapter protein RapZ, SULFATE ION | | Authors: | Gonzalez, G.M, Durica-Mitic, S, Hardwick, S.W, Moncrieffe, M, Resch, M, Neumann, P, Ficner, R, Gorke, B, Luisi, B.F. | | Deposit date: | 2017-06-02 | | Release date: | 2017-08-30 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Structural insights into RapZ-mediated regulation of bacterial amino-sugar metabolism.
Nucleic Acids Res., 45, 2017
|
|
4D8B
 
 | |
5O5S
 
 | | X-ray crystal structure of the RapZ C-terminal domain from Escherichia coli | | Descriptor: | MALONATE ION, RNase adapter protein RapZ | | Authors: | Gonzalez, G.M, Durica-Mitic, S, Hardwick, S.W, Moncrieffe, M, Resch, M, Neumann, P, Ficner, R, Gorke, B, Luisi, B.F. | | Deposit date: | 2017-06-02 | | Release date: | 2017-08-30 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.17 Å) | | Cite: | Structural insights into RapZ-mediated regulation of bacterial amino-sugar metabolism.
Nucleic Acids Res., 45, 2017
|
|
1Y28
 
 | | Crystal structure of the R220A metBJFIXL HEME domain | | Descriptor: | PROTOPORPHYRIN IX CONTAINING FE, Sensor protein fixL | | Authors: | Dunham, C.M, Dioum, E.M, Tuckerman, J.R, Gonzalez, G, Scott, W.G, Gilles-Gonzalez, M.A. | | Deposit date: | 2004-11-21 | | Release date: | 2004-12-07 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A distal arginine in the oxygen-sensing heme-PAS domains is essential to ligand binding, signal transduction, and structure
Biochemistry, 42, 2003
|
|
1DRM
 
 | | CRYSTAL STRUCTURE OF THE LIGAND FREE BJFIXL HEME DOMAIN | | Descriptor: | PROTOPORPHYRIN IX CONTAINING FE, SENSOR PROTEIN FIXL | | Authors: | Gong, W, Hao, B, Mansy, S.S, Gonzalez, G, Gilles-Gonzalez, M.A, Chan, M.K. | | Deposit date: | 2000-01-06 | | Release date: | 2000-01-24 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of a biological oxygen sensor: a new mechanism for heme-driven signal transduction.
Proc.Natl.Acad.Sci.USA, 95, 1998
|
|
3S1I
 
 | | Crystal structure of oxygen-bound hell's gate globin I | | Descriptor: | Hemoglobin-like flavoprotein, OXYGEN MOLECULE, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Teh, A.H, Saito, J.A, Baharuddin, A, Tuckerman, J.R, Newhouse, J.S, Kanbe, M, Newhouse, E.I, Rahim, R.A, Favier, F, Didierjean, C, Sousa, E.H.S, Stott, M.B, Dunfield, P.F, Gonzalez, G, Gilles-Gonzalez, M.A, Najimudin, N, Alam, M. | | Deposit date: | 2011-05-15 | | Release date: | 2011-09-21 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Hell's Gate globin I: an acid and thermostable bacterial hemoglobin resembling mammalian neuroglobin
Febs Lett., 585, 2011
|
|
3S1J
 
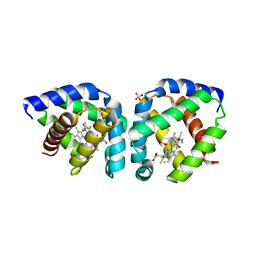 | | Crystal structure of acetate-bound hell's gate globin I | | Descriptor: | ACETATE ION, Hemoglobin-like flavoprotein, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Teh, A.H, Saito, J.A, Baharuddin, A, Tuckerman, J.R, Newhouse, J.S, Kanbe, M, Newhouse, E.I, Rahim, R.A, Favier, F, Didierjean, C, Sousa, E.H.S, Stott, M.B, Dunfield, P.F, Gonzalez, G, Gilles-Gonzalez, M.A, Najimudin, N, Alam, M. | | Deposit date: | 2011-05-15 | | Release date: | 2011-09-21 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Hell's Gate globin I: an acid and thermostable bacterial hemoglobin resembling mammalian neuroglobin
Febs Lett., 585, 2011
|
|
8EO7
 
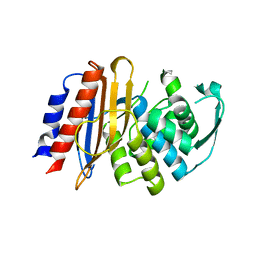 | | Crystal structure of metagenomic beta-lactamase LRA-5 Y69Q/V166E mutant at 2.15 Angstrom resolution | | Descriptor: | beta-lactamase | | Authors: | Power, P, D'Amico Gonzalez, G, Centron, D, Gutkind, G, Handelsman, J, Klinke, S. | | Deposit date: | 2022-10-02 | | Release date: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Playing beta-Lactamase Evolution: Metagenomic Class A beta-Lactamase LRA-5 is an Inactive Enzyme Capable of Rendering an Active beta-Lactamase by Introduction of Y69Q and V166E Substitutions
to be published
|
|
8EO6
 
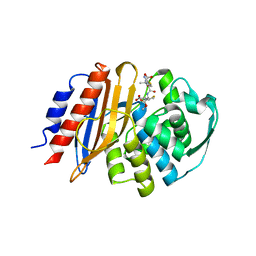 | | Crystal structure of metagenomic class A beta-lactamase precursor LRA-5 in complex with ceftazidime at 2.35 Angstrom resolution | | Descriptor: | ACYLATED CEFTAZIDIME, LRA-5 | | Authors: | Power, P, D'Amico Gonzalez, G, Centron, D, Gutkind, G, Handelsman, J, Klinke, S. | | Deposit date: | 2022-10-02 | | Release date: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Playing beta-Lactamase Evolution: Metagenomic Class A beta-Lactamase LRA-5 is an Inactive Enzyme Capable of Rendering an Active beta-Lactamase by Introduction of Y69Q and V166E Substitutions
to be published
|
|
8EO5
 
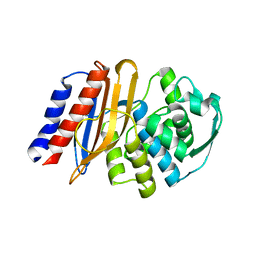 | | Crystal structure of the class A beta-lactamase precursor LRA-5 from an Alaskan soil metagenome at 1.8 Angstrom resolution | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, LRA-5 | | Authors: | Power, P, D'Amico Gonzalez, G, Centron, D, Gutkind, G, Handelsman, J, Klinke, S. | | Deposit date: | 2022-10-02 | | Release date: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Playing beta-Lactamase Evolution: Metagenomic Class A beta-Lactamase LRA-5 is an Inactive Enzyme Capable of Rendering an Active beta-Lactamase by Introduction of Y69Q and V166E Substitutions
to be published
|
|
3SZ7
 
 | |
