1HYW
 
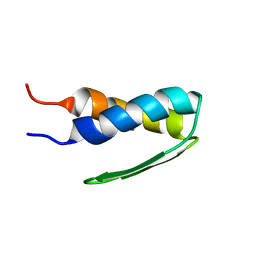 | | SOLUTION STRUCTURE OF BACTERIOPHAGE LAMBDA GPW | | Descriptor: | HEAD-TO-TAIL JOINING PROTEIN W | | Authors: | Maxwell, K.L, Yee, A.A, Booth, V, Arrowsmith, C.H, Gold, M, Davidson, A.R. | | Deposit date: | 2001-01-22 | | Release date: | 2001-04-25 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | The solution structure of bacteriophage lambda protein W, a small morphogenetic protein possessing a novel fold.
J.Mol.Biol., 308, 2001
|
|
1K0H
 
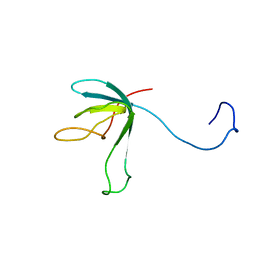 | | Solution structure of bacteriophage lambda gpFII | | Descriptor: | gpFII | | Authors: | Maxwell, K.L, Yee, A.A, Arrowsmith, C.H, Gold, M, Davidson, A.R. | | Deposit date: | 2001-09-19 | | Release date: | 2002-07-17 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | The solution structure of the bacteriophage lambda head-tail joining protein, gpFII.
J.Mol.Biol., 318, 2002
|
|
7B55
 
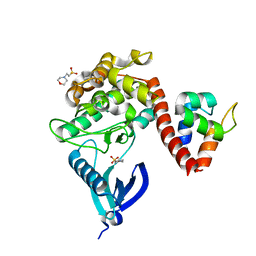 | | Crystal structure of CaMKII-actinin complex bound to MES | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Alpha-actinin-2, Calcium/calmodulin-dependent protein kinase type II subunit alpha | | Authors: | Zhu, J, Gold, M. | | Deposit date: | 2020-12-03 | | Release date: | 2022-06-22 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of CaMKII-actinin complex bound to MES
To Be Published
|
|
7B56
 
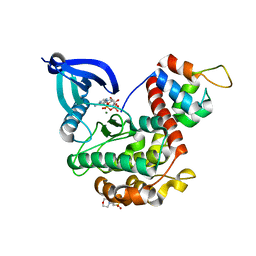 | | Crystal structure of CaMKII-actinin complex bound to AMPPNP | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Alpha-actinin-2, Calcium/calmodulin-dependent protein kinase type II subunit alpha, ... | | Authors: | Zhu, J, Gold, M. | | Deposit date: | 2020-12-03 | | Release date: | 2022-06-22 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Crystal structure of CaMKII-actinin complex bound to MES
To Be Published
|
|
7B57
 
 | | Crystal structure of CaMKII-actinin complex bound to ADP | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ADENOSINE-5'-DIPHOSPHATE, Alpha-actinin-2, ... | | Authors: | Zhu, J, Gold, M. | | Deposit date: | 2020-12-03 | | Release date: | 2022-06-22 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure of CaMKII-actinin complex bound to ADP
To Be Published
|
|
1PL3
 
 | | Cytochrome Domain Of Cellobiose Dehydrogenase, M65H mutant | | Descriptor: | 2-(2-{2-[2-(2-METHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHANOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CADMIUM ION, ... | | Authors: | Rotsaert, F.A.J, Hallberg, B.M, de Vries, S, Moenne-Loccoz, P, Divne, C, Gold, M.H. | | Deposit date: | 2003-06-06 | | Release date: | 2003-07-01 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Biophysical and Structural Analysis of a Novel Heme b Iron Ligation in the Flavocytochrome Cellobiose Dehydrogenase.
J.Biol.Chem., 278, 2003
|
|
6FQJ
 
 | | GluA2(flop) G724C ligand binding core dimer bound to ZK200775 at 2.50 Angstrom resolution | | Descriptor: | Glutamate receptor 2,Glutamate receptor 2, {[7-morpholin-4-yl-2,3-dioxo-6-(trifluoromethyl)-3,4-dihydroquinoxalin-1(2H)-yl]methyl}phosphonic acid | | Authors: | Coombs, I.D, Soto, D, Gold, M.G, Farrant, M.F, Cull-Candy, S.G. | | Deposit date: | 2018-02-14 | | Release date: | 2019-03-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.500017 Å) | | Cite: | Homomeric GluA2(R) AMPA receptors can conduct when desensitized.
Nat Commun, 10, 2019
|
|
6FQG
 
 | | GluA2(flop) G724C ligand binding core dimer bound to L-Glutamate (Form A) at 2.34 Angstrom resolution | | Descriptor: | GLUTAMIC ACID, Glutamate receptor 2 | | Authors: | Coombs, I.D, Soto, D, Gold, M.G, Farrant, M.F, Cull-Candy, S.G. | | Deposit date: | 2018-02-14 | | Release date: | 2019-03-13 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.34139085 Å) | | Cite: | X-ray structure of GluA2 flop G724C ligand binding core dimer bound to glutamate at 2.32 Angstroms resolution
To Be Published
|
|
6FQI
 
 | | GluA2(flop) G724C ligand binding core dimer bound to L-Glutamate (Form B) at 2.91 Angstrom resolution | | Descriptor: | GLUTAMIC ACID, Glutamate receptor 2 | | Authors: | Coombs, I.D, Soto, D, Gold, M.G, Farrant, M.F, Cull-Candy, S.G. | | Deposit date: | 2018-02-14 | | Release date: | 2019-03-13 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.91001153 Å) | | Cite: | Homomeric GluA2(R) AMPA receptors can conduct when desensitized.
Nat Commun, 10, 2019
|
|
6FQK
 
 | | GluA2(flop) S729C ligand binding core dimer bound to ZK200775 at 1.98 Angstrom resolution | | Descriptor: | Glutamate receptor 2,Glutamate receptor 2, {[7-morpholin-4-yl-2,3-dioxo-6-(trifluoromethyl)-3,4-dihydroquinoxalin-1(2H)-yl]methyl}phosphonic acid | | Authors: | Coombs, I.D, Soto, D, Gold, M.G, Farrant, M.F, Cull-Candy, S.G. | | Deposit date: | 2018-02-14 | | Release date: | 2019-03-13 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.98010445 Å) | | Cite: | Homomeric GluA2(R) AMPA receptors can conduct when desensitized.
Nat Commun, 10, 2019
|
|
6FQH
 
 | | GluA2(flop) S729C ligand binding core dimer bound to NBQX at 1.76 Angstrom resolution | | Descriptor: | 6-nitro-2,3-bis(oxidanylidene)-1,4-dihydrobenzo[f]quinoxaline-7-sulfonamide, Glutamate receptor 2,Glutamate receptor 2 | | Authors: | Coombs, I.D, Soto, D, Gold, M.G, Farrant, M.F, Cull-Candy, S.G. | | Deposit date: | 2018-02-14 | | Release date: | 2019-03-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.75940013 Å) | | Cite: | Homomeric GluA2(R) AMPA receptors can conduct when desensitized.
Nat Commun, 10, 2019
|
|
6TS3
 
 | |
5DFS
 
 | | Crystal structure of Spider Monkey Cytochrome C at 1.15 Angstrom | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Cytochrome c, ... | | Authors: | MOU, T.C, McClelland, L.J, Jeakins-Cooley, M.E, Goldes, M.E, SPRANG, S.R, BOWLER, B.E. | | Deposit date: | 2015-08-27 | | Release date: | 2016-03-02 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Disruption of a hydrogen bond network in human versus spider monkey cytochrome c affects heme crevice stability.
J.Inorg.Biochem., 158, 2016
|
|
4WKZ
 
 | | Complex of autonomous ScaG cohesin CohG and X-doc domains | | Descriptor: | ACETATE ION, Autonomous cohesin, CALCIUM ION, ... | | Authors: | Voronov-Goldman, M, Lamed, R, Bayer, E.A, Yaniv, O, Shimon, L.J.W. | | Deposit date: | 2014-10-05 | | Release date: | 2015-12-23 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Complex of autonomous ScaG cohesin CohG and X-doc domains
To Be Published
|
|
5HQ3
 
 | | Stable, high-expression variant of human acetylcholinesterase | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Acetylcholinesterase, O-ETHYLMETHYLPHOSPHONIC ACID ESTER GROUP | | Authors: | Goldenzweig, A, Goldsmith, M, Hill, S.E, Gertman, O, Laurino, P, Ashani, Y, Dym, O, Albeck, S, Unger, T, Prilusky, J, Lieberman, R.L, Aharoni, A, Silman, I, Sussman, J.L, Tawfik, D.S, Fleishman, S.J. | | Deposit date: | 2016-01-21 | | Release date: | 2016-07-27 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Automated Structure- and Sequence-Based Design of Proteins for High Bacterial Expression and Stability.
Mol.Cell, 63, 2016
|
|
7H3H
 
 | | Group deposition for crystallographic fragment screening of Coxsackievirus A16 (G-10) 2A protease -- Crystal structure of Coxsackievirus A16 (G-10) 2A protease in complex with Z57472297 (A71EV2A-x0395) | | Descriptor: | 1-[2-methyl-1,3-bis(oxidanyl)propan-2-yl]-3-phenyl-urea, DIMETHYL SULFOXIDE, Protease 2A, ... | | Authors: | Lithgo, R.M, Fairhead, M, Koekemoer, L, Balcomb, B.H, Capkin, E, Chandran, A.V, Golding, M, Godoy, A.S, Aschenbrenner, J.C, Marples, P.G, Ni, X, Thompson, W, Tomlinson, C.W.E, Wild, C, Winokan, M, Xavier, M.-A.E, Fearon, D, von Delft, F. | | Deposit date: | 2024-04-04 | | Release date: | 2024-04-24 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | Crystallographic Fragment Screen of Coxsackievirus A16 2A Protease identifies new opportunities for the development of broad-spectrum anti-enterovirals.
Biorxiv, 2024
|
|
7H3V
 
 | | Group deposition for crystallographic fragment screening of Coxsackievirus A16 (G-10) 2A protease -- Crystal structure of Coxsackievirus A16 (G-10) 2A protease in complex with Z1198177230 (A71EV2A-x0501) | | Descriptor: | (2M)-2-(2-methoxyphenyl)-1,3,4-oxadiazole, DIMETHYL SULFOXIDE, Protease 2A, ... | | Authors: | Lithgo, R.M, Fairhead, M, Koekemoer, L, Balcomb, B.H, Capkin, E, Chandran, A.V, Golding, M, Godoy, A.S, Aschenbrenner, J.C, Marples, P.G, Ni, X, Thompson, W, Tomlinson, C.W.E, Wild, C, Winokan, M, Xavier, M.-A.E, Fearon, D, von Delft, F. | | Deposit date: | 2024-04-04 | | Release date: | 2024-04-24 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.22 Å) | | Cite: | Crystallographic Fragment Screen of Coxsackievirus A16 2A Protease identifies new opportunities for the development of broad-spectrum anti-enterovirals.
Biorxiv, 2024
|
|
7H39
 
 | | Group deposition for crystallographic fragment screening of Coxsackievirus A16 (G-10) 2A protease -- Crystal structure of Coxsackievirus A16 (G-10) 2A protease in complex with Z50145861 (A71EV2A-x0341) | | Descriptor: | 5-methyl-2-phenyl-2,4-dihydro-3H-pyrazol-3-one, DIMETHYL SULFOXIDE, Protease 2A, ... | | Authors: | Lithgo, R.M, Fairhead, M, Koekemoer, L, Balcomb, B.H, Capkin, E, Chandran, A.V, Golding, M, Godoy, A.S, Aschenbrenner, J.C, Marples, P.G, Ni, X, Thompson, W, Tomlinson, C.W.E, Wild, C, Winokan, M, Xavier, M.-A.E, Fearon, D, von Delft, F. | | Deposit date: | 2024-04-04 | | Release date: | 2024-04-24 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Crystallographic Fragment Screen of Coxsackievirus A16 2A Protease identifies new opportunities for the development of broad-spectrum anti-enterovirals.
Biorxiv, 2024
|
|
7H3Y
 
 | | Group deposition for crystallographic fragment screening of Coxsackievirus A16 (G-10) 2A protease -- Crystal structure of Coxsackievirus A16 (G-10) 2A protease in complex with Z1203107138 (A71EV2A-x0517) | | Descriptor: | 1-ethyl-N-[(4-fluorophenyl)methyl]-1H-pyrazole-4-carboxamide, DIMETHYL SULFOXIDE, Protease 2A, ... | | Authors: | Lithgo, R.M, Fairhead, M, Koekemoer, L, Balcomb, B.H, Capkin, E, Chandran, A.V, Golding, M, Godoy, A.S, Aschenbrenner, J.C, Marples, P.G, Ni, X, Thompson, W, Tomlinson, C.W.E, Wild, C, Winokan, M, Xavier, M.-A.E, Fearon, D, von Delft, F. | | Deposit date: | 2024-04-04 | | Release date: | 2024-04-24 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.11 Å) | | Cite: | Crystallographic Fragment Screen of Coxsackievirus A16 2A Protease identifies new opportunities for the development of broad-spectrum anti-enterovirals.
Biorxiv, 2024
|
|
7H4V
 
 | | Group deposition for crystallographic fragment screening of Coxsackievirus A16 (G-10) 2A protease -- Crystal structure of Coxsackievirus A16 (G-10) 2A protease in complex with Z57663553 (A71EV2A-x1128) | | Descriptor: | 5-phenyl-1,3,4-oxadiazol-2-ol, DIMETHYL SULFOXIDE, Protease 2A, ... | | Authors: | Lithgo, R.M, Fairhead, M, Koekemoer, L, Balcomb, B.H, Capkin, E, Chandran, A.V, Golding, M, Godoy, A.S, Aschenbrenner, J.C, Marples, P.G, Ni, X, Thompson, W, Tomlinson, C.W.E, Wild, C, Winokan, M, Xavier, M.-A.E, Fearon, D, von Delft, F. | | Deposit date: | 2024-04-04 | | Release date: | 2024-04-24 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Crystallographic Fragment Screen of Coxsackievirus A16 2A Protease identifies new opportunities for the development of broad-spectrum anti-enterovirals.
Biorxiv, 2024
|
|
7H2W
 
 | | Group deposition for crystallographic fragment screening of Coxsackievirus A16 (G-10) 2A protease -- Crystal structure of Coxsackievirus A16 (G-10) 2A protease in complex with Z3765495818 (A71EV2A-x0202) | | Descriptor: | (4R)-imidazo[1,2-b]pyridazine-3-sulfonamide, DIMETHYL SULFOXIDE, Protease 2A, ... | | Authors: | Lithgo, R.M, Fairhead, M, Koekemoer, L, Balcomb, B.H, Capkin, E, Chandran, A.V, Golding, M, Godoy, A.S, Aschenbrenner, J.C, Marples, P.G, Ni, X, Thompson, W, Tomlinson, C.W.E, Wild, C, Winokan, M, Xavier, M.-A.E, Fearon, D, von Delft, F. | | Deposit date: | 2024-04-04 | | Release date: | 2024-04-24 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Crystallographic Fragment Screen of Coxsackievirus A16 2A Protease identifies new opportunities for the development of broad-spectrum anti-enterovirals.
Biorxiv, 2024
|
|
7H36
 
 | | Group deposition for crystallographic fragment screening of Coxsackievirus A16 (G-10) 2A protease -- Crystal structure of Coxsackievirus A16 (G-10) 2A protease in complex with Z1688504114 (A71EV2A-x0310) | | Descriptor: | 2-cyano-~{N}-(1,3,5-trimethylpyrazol-4-yl)ethanamide, DIMETHYL SULFOXIDE, Protease 2A, ... | | Authors: | Lithgo, R.M, Fairhead, M, Koekemoer, L, Balcomb, B.H, Capkin, E, Chandran, A.V, Golding, M, Godoy, A.S, Aschenbrenner, J.C, Marples, P.G, Ni, X, Thompson, W, Tomlinson, C.W.E, Wild, C, Winokan, M, Xavier, M.-A.E, Fearon, D, von Delft, F. | | Deposit date: | 2024-04-04 | | Release date: | 2024-04-24 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | Crystallographic Fragment Screen of Coxsackievirus A16 2A Protease identifies new opportunities for the development of broad-spectrum anti-enterovirals.
Biorxiv, 2024
|
|
7H3E
 
 | | Group deposition for crystallographic fragment screening of Coxsackievirus A16 (G-10) 2A protease -- Crystal structure of Coxsackievirus A16 (G-10) 2A protease in complex with Z56921372 (A71EV2A-x0375) | | Descriptor: | DIMETHYL SULFOXIDE, N-(2-methoxyphenyl)thiourea, Protease 2A, ... | | Authors: | Lithgo, R.M, Fairhead, M, Koekemoer, L, Balcomb, B.H, Capkin, E, Chandran, A.V, Golding, M, Godoy, A.S, Aschenbrenner, J.C, Marples, P.G, Ni, X, Thompson, W, Tomlinson, C.W.E, Wild, C, Winokan, M, Xavier, M.-A.E, Fearon, D, von Delft, F. | | Deposit date: | 2024-04-04 | | Release date: | 2024-04-24 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | Crystallographic Fragment Screen of Coxsackievirus A16 2A Protease identifies new opportunities for the development of broad-spectrum anti-enterovirals.
Biorxiv, 2024
|
|
7H3U
 
 | | Group deposition for crystallographic fragment screening of Coxsackievirus A16 (G-10) 2A protease -- Crystal structure of Coxsackievirus A16 (G-10) 2A protease in complex with Z1198158918 (A71EV2A-x0497) | | Descriptor: | 1H-indazole-3-carbonitrile, DIMETHYL SULFOXIDE, Protease 2A, ... | | Authors: | Lithgo, R.M, Fairhead, M, Koekemoer, L, Balcomb, B.H, Capkin, E, Chandran, A.V, Golding, M, Godoy, A.S, Aschenbrenner, J.C, Marples, P.G, Ni, X, Thompson, W, Tomlinson, C.W.E, Wild, C, Winokan, M, Xavier, M.-A.E, Fearon, D, von Delft, F. | | Deposit date: | 2024-04-04 | | Release date: | 2024-04-24 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.31 Å) | | Cite: | Crystallographic Fragment Screen of Coxsackievirus A16 2A Protease identifies new opportunities for the development of broad-spectrum anti-enterovirals.
Biorxiv, 2024
|
|
7H2U
 
 | | Group deposition for crystallographic fragment screening of Coxsackievirus A16 (G-10) 2A protease -- Crystal structure of Coxsackievirus A16 (G-10) 2A protease in complex with Z355202286 (A71EV2A-x0188) | | Descriptor: | (2R)-2-ethyl-N,N-dimethylmorpholine-4-sulfonamide, DIMETHYL SULFOXIDE, Protease 2A, ... | | Authors: | Lithgo, R.M, Fairhead, M, Koekemoer, L, Balcomb, B.H, Capkin, E, Chandran, A.V, Golding, M, Godoy, A.S, Aschenbrenner, J.C, Marples, P.G, Ni, X, Thompson, W, Tomlinson, C.W.E, Wild, C, Winokan, M, Xavier, M.-A.E, Fearon, D, von Delft, F. | | Deposit date: | 2024-04-04 | | Release date: | 2024-04-24 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.11 Å) | | Cite: | Crystallographic Fragment Screen of Coxsackievirus A16 2A Protease identifies new opportunities for the development of broad-spectrum anti-enterovirals.
Biorxiv, 2024
|
|
