3L7F
 
 | | Structure of IL-13 antibody H2L6, A humanized variant OF C836 | | Descriptor: | CALCIUM ION, H2L6 HEAVY CHAIN, H2L6 LIGHT CHAIN, ... | | Authors: | Teplyakov, A, Obmolova, G, Malia, T, Gilliland, G.L. | | Deposit date: | 2009-12-28 | | Release date: | 2010-11-10 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Human framework adaptation of a mouse anti-human IL-13 antibody.
J.Mol.Biol., 398, 2010
|
|
3IFD
 
 | |
3I84
 
 | |
2IMD
 
 | | Structure of SeMet 2-hydroxychromene-2-carboxylate isomerase (HCCA isomerase) | | Descriptor: | (2S)-2-HYDROXY-2H-CHROMENE-2-CARBOXYLIC ACID, (3E)-4-(2-HYDROXYPHENYL)-2-OXOBUT-3-ENOIC ACID, 2-hydroxychromene-2-carboxylate isomerase, ... | | Authors: | Thompson, L.C, Ladner, J.E, Codreanu, S.G, Harp, J, Gilliland, G.L, Armstrong, R.N. | | Deposit date: | 2006-10-04 | | Release date: | 2007-06-12 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | 2-Hydroxychromene-2-carboxylic Acid Isomerase: A Kappa Class Glutathione Transferase from Pseudomonas putida
Biochemistry, 46, 2007
|
|
2IMF
 
 | | 2-Hydroxychromene-2-carboxylate Isomerase: a Kappa Class Glutathione-S-Transferase from Pseudomonas putida | | Descriptor: | 2-hydroxychromene-2-carboxylate isomerase, 3-CYCLOHEXYL-1-PROPYLSULFONIC ACID, 4-(2-METHOXYPHENYL)-2-OXOBUT-3-ENOIC ACID, ... | | Authors: | Thompson, L.C, Ladner, J.E, Codreanu, S.G, Harp, J, Gilliland, G.L, Armstrong, R.N. | | Deposit date: | 2006-10-04 | | Release date: | 2007-06-12 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | 2-Hydroxychromene-2-carboxylic acid isomerase: a kappa class glutathione transferase from Pseudomonas putida.
Biochemistry, 46, 2007
|
|
3I85
 
 | |
2IME
 
 | | 2-Hydroxychromene-2-carboxylate Isomerase: a Kappa Class Glutathione-S-Transferase from Pseudomonas putida | | Descriptor: | (2S)-2-HYDROXY-2H-CHROMENE-2-CARBOXYLIC ACID, (3E)-4-(2-HYDROXYPHENYL)-2-OXOBUT-3-ENOIC ACID, 2-hydroxychromene-2-carboxylate isomerase, ... | | Authors: | Thompson, L.C, Ladner, J.E, Codreanu, S.G, Harp, J, Gilliland, G.L, Armstrong, R.N. | | Deposit date: | 2006-10-04 | | Release date: | 2007-06-12 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | 2-Hydroxychromene-2-carboxylic Acid Isomerase: A Kappa Class Glutathione Transferase from Pseudomonas putida
Biochemistry, 46, 2007
|
|
3I2C
 
 | | Crystal structure of anti-IL-23 antibody CNTO4088 | | Descriptor: | CNTO4088 HEAVY CHAIN, CNTO4088 LIGHT CHAIN, GLYCEROL | | Authors: | Teplyakov, A, Obmolova, G, Gilliland, G.L. | | Deposit date: | 2009-06-29 | | Release date: | 2009-07-07 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Noncanonical conformation of CDR L1 in the anti-IL-23 antibody CNTO4088.
Acta Crystallogr.,Sect.F, 66, 2010
|
|
1YI7
 
 | | Beta-d-xylosidase (selenomethionine) XYND from Clostridium Acetobutylicum | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Beta-xylosidase, family 43 glycosyl hydrolase, ... | | Authors: | Teplyakov, A, Fedorov, E, Gilliland, G.L, Almo, S.C, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2005-01-11 | | Release date: | 2005-02-22 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of beta-xylosidase from clostridium acetobutylicum
To be Published
|
|
1Y7B
 
 | | BETA-D-XYLOSIDASE, A FAMILY 43 GLYCOSIDE HYDROLASE | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Beta-xylosidase, family 43 glycosyl hydrolase, ... | | Authors: | Teplyakov, A, Fedorov, E, Gilliland, G.L, Almo, S.C, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2004-12-08 | | Release date: | 2005-01-25 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of beta-xylosidase from Clostridium acetobutylicum
To be Published
|
|
3G6D
 
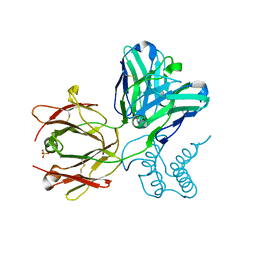 | | Crystal structure of the complex between CNTO607 Fab and IL-13 | | Descriptor: | CNTO607 Fab Heavy chain, CNTO607 Fab Light chain, Interleukin-13, ... | | Authors: | Teplyakov, A, Obmolova, G, Gilliland, G.L. | | Deposit date: | 2009-02-06 | | Release date: | 2009-04-07 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Epitope mapping of anti-interleukin-13 neutralizing antibody CNTO607.
J.Mol.Biol., 389, 2009
|
|
1WSA
 
 | | STRUCTURE OF L-ASPARAGINASE II PRECURSOR | | Descriptor: | ASPARAGINE AMIDOHYDROLASE | | Authors: | Lubkowski, J, Palm, G.J, Gilliland, G.L, Derst, C, Rohm, K.-H, Wlodawer, A. | | Deposit date: | 1996-08-15 | | Release date: | 1997-04-01 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure and amino acid sequence of Wolinella succinogenes L-asparaginase.
Eur.J.Biochem., 241, 1996
|
|
4I1B
 
 | | FUNCTIONAL IMPLICATIONS OF INTERLEUKIN-1BETA BASED ON THE THREE-DIMENSIONAL STRUCTURE | | Descriptor: | INTERLEUKIN-1 BETA | | Authors: | Veerapandian, B, Poulos, T.L, Gilliland, G.L, Raag, R, Svensson, L.A, Masui, Y, Hirai, Y. | | Deposit date: | 1990-03-27 | | Release date: | 1990-04-15 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Functional implications of interleukin-1 beta based on the three-dimensional structure.
Proteins, 12, 1992
|
|
1CLS
 
 | | CROSS-LINKED HUMAN HEMOGLOBIN DEOXY | | Descriptor: | HEMOGLOBIN, OXYGEN ATOM, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Ji, X, Fronticelli, C, Bucci, E, Gilliland, G.L. | | Deposit date: | 1995-08-29 | | Release date: | 1996-10-14 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Positive and negative cooperativities at subsequent steps of oxygenation regulate the allosteric behavior of multistate sebacylhemoglobin.
Biochemistry, 35, 1996
|
|
1HW5
 
 | | THE CAP/CRP VARIANT T127L/S128A | | Descriptor: | ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE, CATABOLITE GENE ACTIVATOR | | Authors: | Chu, S.Y, Tordova, M, Gilliland, G.L, Gorshkova, I, Shi, Y. | | Deposit date: | 2001-01-09 | | Release date: | 2001-01-17 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | The structure of the T127L/S128A mutant of cAMP receptor protein facilitates promoter site binding
J.Biol.Chem., 276, 2001
|
|
1SPB
 
 | | SUBTILISIN BPN' PROSEGMENT (77 RESIDUES) COMPLEXED WITH A MUTANT SUBTILISIN BPN' (266 RESIDUES). CRYSTAL PH 4.6. CRYSTALLIZATION TEMPERATURE 20 C DIFFRACTION TEMPERATURE-160 C | | Descriptor: | SODIUM ION, SUBTILISIN BPN', SUBTILISIN BPN' PROSEGMENT | | Authors: | Gallagher, D.T, Gilliland, G.L, Wang, L, Bryan, P.N. | | Deposit date: | 1995-06-21 | | Release date: | 1995-10-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The prosegment-subtilisin BPN' complex: crystal structure of a specific 'foldase'.
Structure, 3, 1995
|
|
1TDJ
 
 | | THREONINE DEAMINASE (BIOSYNTHETIC) FROM E. COLI | | Descriptor: | BIOSYNTHETIC THREONINE DEAMINASE, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Gallagher, D.T, Gilliland, G.L, Xiao, G, Eisenstein, E. | | Deposit date: | 1998-03-27 | | Release date: | 1998-10-14 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure and control of pyridoxal phosphate dependent allosteric threonine deaminase.
Structure, 6, 1998
|
|
1BHN
 
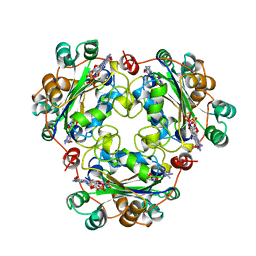 | | NUCLEOSIDE DIPHOSPHATE KINASE ISOFORM A FROM BOVINE RETINA | | Descriptor: | GUANOSINE-3',5'-MONOPHOSPHATE, GUANOSINE-5'-DIPHOSPHATE, NUCLEOSIDE DIPHOSPHATE TRANSFERASE | | Authors: | Ladner, J.E, Abdulaev, N.G, Kakuev, D.L, Karaschuk, G.N, Tordova, M, Eisenstein, E, Fujiwara, J.H, Ridge, K.D, Gilliland, G.L. | | Deposit date: | 1998-06-10 | | Release date: | 1999-02-16 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The three-dimensional structures of two isoforms of nucleoside diphosphate kinase from bovine retina.
Acta Crystallogr.,Sect.D, 55, 1999
|
|
1BE4
 
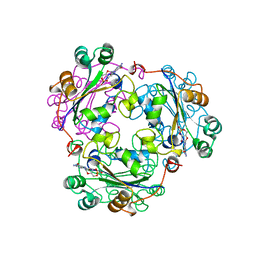 | | NUCLEOSIDE DIPHOSPHATE KINASE ISOFORM B FROM BOVINE RETINA | | Descriptor: | CYCLIC GUANOSINE MONOPHOSPHATE, NUCLEOSIDE DIPHOSPHATE TRANSFERASE | | Authors: | Ladner, J.E, Abdulaev, N.G, Kakuev, D.L, Karaschuk, G.N, Tordova, M, Eisenstein, E, Fujiwara, J.H, Ridge, K.D, Gilliland, G.L. | | Deposit date: | 1998-05-19 | | Release date: | 1999-01-13 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Nucleoside diphosphate kinase from bovine retina: purification, subcellular localization, molecular cloning, and three-dimensional structure.
Biochemistry, 37, 1998
|
|
3NCJ
 
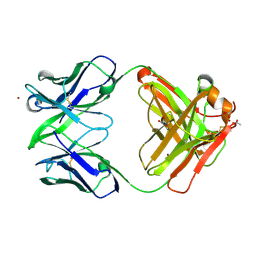 | | Crystal structure of Fab15 Mut8 | | Descriptor: | ACETATE ION, Fab15 Mut8 heavy chain, Fab15 Mut8 light chain, ... | | Authors: | Luo, J, Feng, Y, Gilliland, G.L. | | Deposit date: | 2010-06-04 | | Release date: | 2010-08-18 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Co-evolution of antibody stability and Vk CDR-L3 canonical structure
To be Published
|
|
3NAA
 
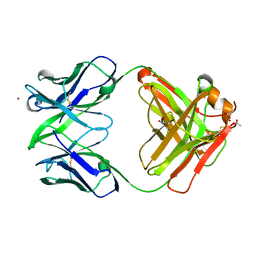 | | Crystal structure of Fab15 Mut5 | | Descriptor: | ACETATE ION, Fab15 Mut5 heavy chain, Fab15 Mut5 light chain, ... | | Authors: | Luo, J, Feng, Y, Gilliland, G.L. | | Deposit date: | 2010-06-01 | | Release date: | 2010-08-18 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Co-evolution of antibody stability and Vk CDR-L3 canonical structure
To be Published
|
|
3MBX
 
 | | Crystal structure of chimeric antibody X836 | | Descriptor: | PHOSPHATE ION, SODIUM ION, X836 HEAVY CHAIN, ... | | Authors: | Teplyakov, A, Obmolova, G, Gilliland, G.L. | | Deposit date: | 2010-03-26 | | Release date: | 2010-06-30 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | On the domain pairing in chimeric antibodies.
Mol.Immunol., 47, 2010
|
|
3MCK
 
 | |
1FLZ
 
 | | URACIL DNA GLYCOSYLASE WITH UAAP | | Descriptor: | URACIL, URACIL-DNA GLYCOSYLASE | | Authors: | Werner, R.M, Jiang, Y.L, Gordley, R.G, Jagadeesh, G.J, Ladner, J.E, Xiao, G, Tordova, M, Gilliland, G.L, Stivers, J.T. | | Deposit date: | 2000-08-15 | | Release date: | 2001-01-17 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Stressing-out DNA? The contribution of serine-phosphodiester interactions in catalysis by uracil DNA glycosylase.
Biochemistry, 39, 2000
|
|
3MCL
 
 | |
