8EY8
 
 | |
8EY6
 
 | |
8EY7
 
 | |
3BXY
 
 | | Crystal structure of tetrahydrodipicolinate N-succinyltransferase from E. coli | | Descriptor: | 2,3,4,5-tetrahydropyridine-2,6-dicarboxylate N-succinyltransferase | | Authors: | Kozlov, G, Gehring, K, Montreal-Kingston Bacterial Structural Genomics Initiative (BSGI) | | Deposit date: | 2008-01-15 | | Release date: | 2008-01-29 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of Escherichia coli tetrahydrodipicolinate N-succinyltransferase reveals the role of a conserved C-terminal helix in cooperative substrate binding.
Febs Lett., 582, 2008
|
|
3BCY
 
 | | Crystal structure of YER067W | | Descriptor: | Protein YER067W | | Authors: | Kozlov, G, Gehring, K. | | Deposit date: | 2007-11-13 | | Release date: | 2008-11-18 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural and functional study of YER067W, a new protein involved in yeast metabolism control and drug resistance.
Plos One, 5, 2010
|
|
3DF0
 
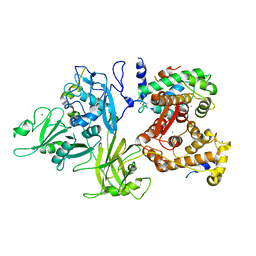 | | Calcium-dependent complex between m-calpain and calpastatin | | Descriptor: | CALCIUM ION, Calpain small subunit 1, Calpain-2 catalytic subunit, ... | | Authors: | Moldoveanu, T, Gehring, K, Green, D.R. | | Deposit date: | 2008-06-11 | | Release date: | 2008-11-11 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Concerted multi-pronged attack by calpastatin to occlude the catalytic cleft of heterodimeric calpains.
Nature, 456, 2008
|
|
2X5N
 
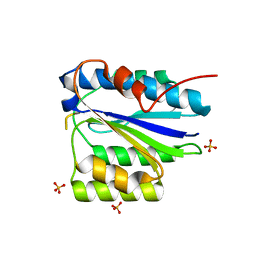 | | Crystal Structure of the SpRpn10 VWA domain | | Descriptor: | 26S PROTEASOME REGULATORY SUBUNIT RPN10, SULFATE ION | | Authors: | Riedinger, C, Boehringer, J, Trempe, J.-F, Lowe, E.D, Brown, N.R, Gehring, K, Noble, M.E.M, Gordon, C, Endicott, J.A. | | Deposit date: | 2010-02-10 | | Release date: | 2010-08-25 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | The Structure of Rpn10 and its Interactions with Polyubiquitin Chains and the Proteasome Subunit Rpn12.
J.Biol.Chem., 285, 2010
|
|
7SOR
 
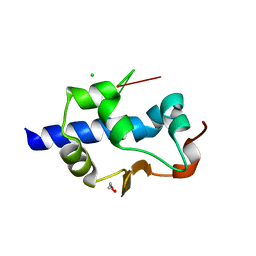 | | LaM domain of human LARP1 in complex with AAA RNA | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Isoform 2 of La-related protein 1, ... | | Authors: | Kozlov, G, Gehring, K. | | Deposit date: | 2021-11-01 | | Release date: | 2022-08-03 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structural basis of 3'-end poly(A) RNA recognition by LARP1.
Nucleic Acids Res., 50, 2022
|
|
7SOS
 
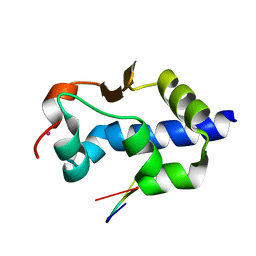 | | LaM domain of human LARP1 in complex with AAAA RNA | | Descriptor: | Isoform 2 of La-related protein 1, POTASSIUM ION, RNA (5'-R(*AP*AP*AP*A)-3') | | Authors: | Kozlov, G, Gehring, K. | | Deposit date: | 2021-11-01 | | Release date: | 2022-08-03 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Structural basis of 3'-end poly(A) RNA recognition by LARP1.
Nucleic Acids Res., 50, 2022
|
|
7US1
 
 | | Structure of parkin (R0RB) bound to two phospho-ubiquitin molecules | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, E3 ubiquitin-protein ligase parkin, ... | | Authors: | Fakih, R, Sauve, V, Gehring, K. | | Deposit date: | 2022-04-22 | | Release date: | 2022-06-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.484 Å) | | Cite: | Structure of the second phosphoubiquitin-binding site in parkin.
J.Biol.Chem., 298, 2022
|
|
3RG0
 
 | | Structural and functional relationships between the lectin and arm domains of calreticulin | | Descriptor: | CALCIUM ION, Calreticulin | | Authors: | Kozlov, G, Pocanschi, C.L, Brockmeier, U, Williams, D.B, Gehring, K. | | Deposit date: | 2011-04-07 | | Release date: | 2011-06-01 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | Structural and Functional Relationships between the Lectin and Arm Domains of Calreticulin.
J.Biol.Chem., 286, 2011
|
|
6N7E
 
 | |
3UVT
 
 | |
4F02
 
 | |
5BU2
 
 | | Structure of the C-terminal domain of lpg1496 from Legionella pneumophila in complex with nucleotide | | Descriptor: | ADENOSINE MONOPHOSPHATE, ADENOSINE-5'-DIPHOSPHATE, alpha-D-ribofuranose, ... | | Authors: | Wong, K, Kozlov, G, Gehring, K, Montreal-Kingston Bacterial Structural Genomics Initiative (BSGI) | | Deposit date: | 2015-06-03 | | Release date: | 2015-08-26 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Structure of the Legionella Effector, lpg1496, Suggests a Role in Nucleotide Metabolism.
J.Biol.Chem., 290, 2015
|
|
5KES
 
 | | Solution structure of the yeast Ddi1 HDD domain | | Descriptor: | DNA damage-inducible protein 1 | | Authors: | Trempe, J.-F, Ratcliffe, C, Veverka, V, Saskova, K, Gehring, K. | | Deposit date: | 2016-06-10 | | Release date: | 2016-10-05 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural studies of the yeast DNA damage-inducible protein Ddi1 reveal domain architecture of this eukaryotic protein family.
Sci Rep, 6, 2016
|
|
2QHO
 
 | |
5BTW
 
 | | Structure of the N-terminal domain of lpg1496 from Legionella pneumophila | | Descriptor: | Uncharacterized protein | | Authors: | Wong, K, Kozlov, G, Gehring, K, Montreal-Kingston Bacterial Structural Genomics Initiative (BSGI) | | Deposit date: | 2015-06-03 | | Release date: | 2015-08-26 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Structure of the Legionella Effector, lpg1496, Suggests a Role in Nucleotide Metabolism.
J.Biol.Chem., 290, 2015
|
|
5BU1
 
 | | Structure of the truncated C-terminal domain of lpg1496 from Legionella pneumophila | | Descriptor: | LPG1496, MALONATE ION | | Authors: | Wong, K, Kozlov, G, Gehring, K, Montreal-Kingston Bacterial Structural Genomics Initiative (BSGI) | | Deposit date: | 2015-06-03 | | Release date: | 2015-08-26 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure of the Legionella Effector, lpg1496, Suggests a Role in Nucleotide Metabolism.
J.Biol.Chem., 290, 2015
|
|
5K35
 
 | | Structure of the Legionella effector, AnkB, in complex with human Skp1 | | Descriptor: | Ankyrin-repeat protein B, S-phase kinase-associated protein 1 | | Authors: | Wong, K, Kozlov, G, Gehring, K, Montreal-Kingston Bacterial Structural Genomics Initiative (BSGI) | | Deposit date: | 2016-05-19 | | Release date: | 2017-01-25 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structural Mimicry by a Bacterial F Box Effector Hijacks the Host Ubiquitin-Proteasome System.
Structure, 25, 2017
|
|
1ME0
 
 | | Chimeric hairpin with 2',5'-linked RNA loop and DNA stem | | Descriptor: | DNA/RNA (5'-D(*GP*GP*AP*C)-R(P*(U25)P*(U25)P*(C25)P*(5GP))-D(P*GP*TP*CP*C)-3') | | Authors: | Denisov, A.Y, Hannoush, R.N, Gehring, K, Damha, M.J. | | Deposit date: | 2002-08-07 | | Release date: | 2003-08-19 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | A Novel RNA Motif Based on the Structure of Unusually Stable 2',5'-Linked r(UUCG) Loops
J.AM.CHEM.SOC., 125, 2003
|
|
5K34
 
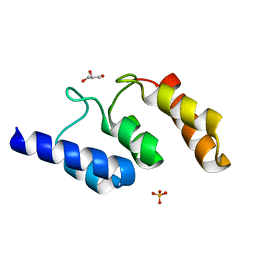 | | Structure of the ankyrin domain of AnkB from Legionella Pneumophila | | Descriptor: | Ankyrin-repeat protein B, GLYCEROL, SULFATE ION | | Authors: | Wong, K, Kozlov, G, Gehring, K, Montreal-Kingston Bacterial Structural Genomics Initiative (BSGI) | | Deposit date: | 2016-05-19 | | Release date: | 2017-01-25 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Structural Mimicry by a Bacterial F Box Effector Hijacks the Host Ubiquitin-Proteasome System.
Structure, 25, 2017
|
|
5BTX
 
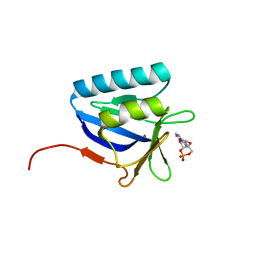 | | Structure of the N-terminal domain of lpg1496 from Legionella pneumophila in complex with nucleotide | | Descriptor: | ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE, lpg1496 | | Authors: | Wong, K, Kozlov, G, Gehring, K, Montreal-Kingston Bacterial Structural Genomics Initiative (BSGI) | | Deposit date: | 2015-06-03 | | Release date: | 2015-08-26 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of the Legionella Effector, lpg1496, Suggests a Role in Nucleotide Metabolism.
J.Biol.Chem., 290, 2015
|
|
5BU0
 
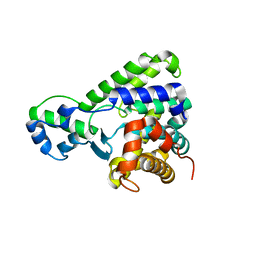 | | Structure of the C-terminal domain of lpg1496 from Legionella pneumophila | | Descriptor: | lpg1496 | | Authors: | Wong, K, Kozlov, G, Gehring, K, Montreal-Kingston Bacterial Structural Genomics Initiative (BSGI) | | Deposit date: | 2015-06-03 | | Release date: | 2015-08-26 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structure of the Legionella Effector, lpg1496, Suggests a Role in Nucleotide Metabolism.
J.Biol.Chem., 290, 2015
|
|
5BTZ
 
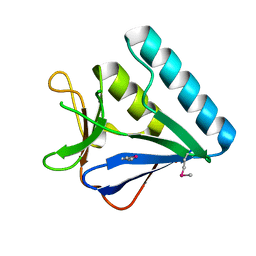 | | Structure of the middle domain of lpg1496 from Legionella pneumophila in P212121 space group | | Descriptor: | lpg1496 | | Authors: | Wong, K, Kozlov, G, Gehring, K, Montreal-Kingston Bacterial Structural Genomics Initiative (BSGI) | | Deposit date: | 2015-06-03 | | Release date: | 2015-08-26 | | Last modified: | 2020-01-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure of the Legionella Effector, lpg1496, Suggests a Role in Nucleotide Metabolism.
J.Biol.Chem., 290, 2015
|
|
