5LTV
 
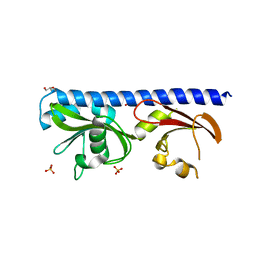 | | LIGAND BINDING DOMAIN OF PSEUDOMONAS AERUGINOSA PAO1 AMINO ACID CHEMORECPETOR PCTC IN COMPLEX WITH GABA | | Descriptor: | ACETATE ION, Chemotactic transducer PctC, GAMMA-AMINO-BUTANOIC ACID, ... | | Authors: | Gavira, J.A, Rico-Jimenez, M, Conejero-Muriel, M, Krell, T. | | Deposit date: | 2016-09-07 | | Release date: | 2017-09-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | How Bacterial Chemoreceptors Evolve Novel Ligand Specificities
Mbio, 2020
|
|
5LTX
 
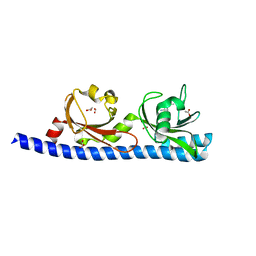 | | LIGAND BINDING DOMAIN OF PSEUDOMONAS AERUGINOSA PAO1 AMINO ACID CHEMORECEPTOR PCTA IN COMPLEX WITH L-MET | | Descriptor: | ACETATE ION, Chemotaxis protein, FORMIC ACID, ... | | Authors: | Gavira, J.A, Rico-Gimenez, M, Ortega, A, Conejero-Muriel, M, Zhulin, I, Krell, T. | | Deposit date: | 2016-09-07 | | Release date: | 2017-09-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | How Bacterial Chemoreceptors Evolve Novel Ligand Specificities
Mbio, 2020
|
|
4C6Y
 
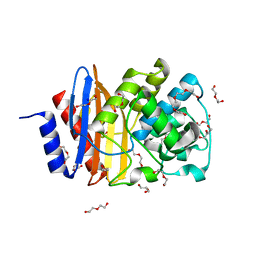 | | Ancestral PNCA (last common ancestors of Gram-positive and Gram- negative bacteria) beta-lactamase class A | | Descriptor: | ACETATE ION, BETA-LACTAMASE, CHLORIDE ION, ... | | Authors: | Gavira, J.A, Risso, V.A, Sanchez-Ruiz, J.M. | | Deposit date: | 2013-09-19 | | Release date: | 2014-04-16 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.799 Å) | | Cite: | Phenotypic Comparisons of Consensus Variants Versus Laboratory Resurrections of Precambrian Proteins.
Proteins, 82, 2014
|
|
4C75
 
 | | Consensus (ALL-CON) beta-lactamase class A | | Descriptor: | ACETATE ION, BETA-LACTAMASE, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Gavira, J.A, Risso, V.A, Sanchez-Ruiz, J.M. | | Deposit date: | 2013-09-19 | | Release date: | 2014-04-16 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Phenotypic Comparisons of Consensus Variants Versus Laboratory Resurrections of Precambrian Proteins.
Proteins, 82, 2014
|
|
7PTH
 
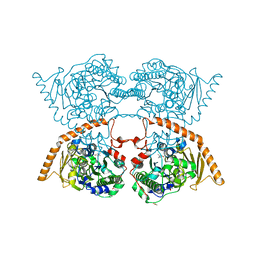 | |
7PTJ
 
 | |
7Q1L
 
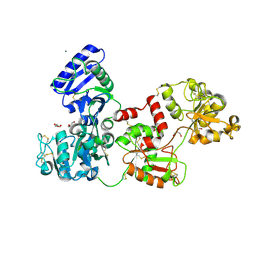 | | Glycosilated Human Serum Apo-tranferrin | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, ... | | Authors: | Gavira, J.A, Moreno, A, Campos-Escamilla, C, Gonzalez-Ramirez, L.A, Siliqi, D. | | Deposit date: | 2021-10-20 | | Release date: | 2022-03-16 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | X-ray Characterization of Conformational Changes of Human Apo- and Holo-Transferrin.
Int J Mol Sci, 22, 2021
|
|
6Z1M
 
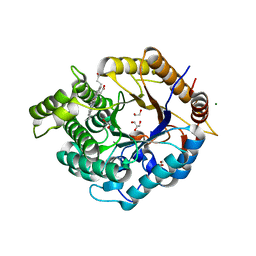 | | Structure of an Ancestral glycosidase (family 1) bound to heme | | Descriptor: | 1,2-ETHANEDIOL, Ancestral reconstructed glycosidase, GLYCEROL, ... | | Authors: | Gavira, J.A, Risso, V.A, Sanchez-Ruiz, J.M, Gamiz-Arco, G, Gutierrez-Rus, L, Ibarra-Molero, B, Oshino, Y, Petrovic, D, Romero-Rivera, A, Seelig, B, Kamerlin, S.C.L, Gaucher, E.A. | | Deposit date: | 2020-05-14 | | Release date: | 2020-07-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Heme-binding enables allosteric modulation in an ancient TIM-barrel glycosidase.
Nat Commun, 12, 2021
|
|
7QEK
 
 | | Structure of the ligand binding domain of the antibiotic biosynthesis regulator AdmX from the rhizobacterium Serratia plymuthica A153 bound to the auxin indole-3-piruvic acid (IPA). | | Descriptor: | 3-(1H-INDOL-3-YL)-2-OXOPROPANOIC ACID, MAGNESIUM ION, regulator AdmX | | Authors: | Gavira, J.A, Rico-Jimenez, M, Castellvi, A, Krell, T, Matilla, M.A. | | Deposit date: | 2021-12-03 | | Release date: | 2022-12-14 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Emergence of an Auxin Sensing Domain in Plant-Associated Bacteria.
Mbio, 14, 2023
|
|
7QEJ
 
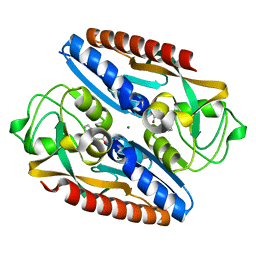 | | Structure of the ligand binding domain of the antibiotic biosynthesis regulator AdmX from the rhizobacterium Serratia plymuthica A153 bound to the auxin indole-3-acetic acid (IAA). | | Descriptor: | 1H-INDOL-3-YLACETIC ACID, MAGNESIUM ION, TRANSCRIPTIONAL REGULATOR AdmX | | Authors: | Gavira, J.A, Rico-Jimenez, M, Castellvi, A, Krell, T, Matilla, M.A. | | Deposit date: | 2021-12-03 | | Release date: | 2022-12-14 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Emergence of an Auxin Sensing Domain in Plant-Associated Bacteria.
Mbio, 14, 2023
|
|
2YFB
 
 | | X-ray structure of McpS ligand binding domain in complex with succinate | | Descriptor: | ACETATE ION, METHYL-ACCEPTING CHEMOTAXIS TRANSDUCER, SUCCINIC ACID, ... | | Authors: | Gavira, J.A, Pineda-Molina, E, Krell, T. | | Deposit date: | 2011-04-05 | | Release date: | 2012-04-18 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Evidence for Chemoreceptors with Bimodular Ligand-Binding Regions Harboring Two Signal-Binding Sites.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
6YBF
 
 | |
3DC8
 
 | |
1Z2U
 
 | | The 1.1A crystallographic structure of ubiquitin-conjugating enzyme (ubc-2) from Caenorhabditis elegans: functional and evolutionary significance | | Descriptor: | (R,R)-2,3-BUTANEDIOL, CHLORIDE ION, SODIUM ION, ... | | Authors: | Gavira, J.A, DiGiamamarino, E, Tempel, W, Liu, Z.J, Wang, B.C, Meehan, E, Ng, J.D, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2005-03-09 | | Release date: | 2005-03-22 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | The 1.1A crystallographic structure of ubiquitin-conjugating enzyme (ubc-2) from Caenorhabditis elegans: functional and evolutionary significance
To be published
|
|
1Z3D
 
 | | Protein crystal growth improvement leading to the 2.5A crystallographic structure of ubiquitin-conjugating enzyme (ubc-1) from Caenorhabditis elegans | | Descriptor: | Ubiquitin-conjugating enzyme E2 1 | | Authors: | Gavira, J.A, DiGiammarino, E, Tempel, W, Toh, D, Liu, Z.J, Wang, B.C, Meehan, E, Ng, J.D, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2005-03-11 | | Release date: | 2005-03-22 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Protein crystal growth improvement leading to the 2.5A crystallographic structure of ubiquitin-conjugating enzyme (ubc-1) from Caenorhabditis elegans
To be Published
|
|
8BMV
 
 | | Ligand binding domain of the P. Putida receptor McpH in complex with Uric acid | | Descriptor: | Methyl-accepting chemotaxis protein McpH, URIC ACID | | Authors: | Gavira, J.A, Krell, T, Fernandez, M, Martinez-Rodriguez, S. | | Deposit date: | 2022-11-11 | | Release date: | 2024-07-24 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Ubiquitous purine sensor modulates diverse signal transduction pathways in bacteria.
Nat Commun, 15, 2024
|
|
1ZZY
 
 | |
6S33
 
 | | Ligand binding domain of the P. putida receptor PcaY_PP in complex with Protocatechuate | | Descriptor: | 3,4-DIHYDROXYBENZOIC ACID, ACETATE ION, Aromatic acid chemoreceptor | | Authors: | Gavira, J.A, Mantilla, M.A, Fernandez, M, Krell, T. | | Deposit date: | 2019-06-24 | | Release date: | 2020-10-21 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | The structural basis for signal promiscuity in a bacterial chemoreceptor.
Febs J., 288, 2021
|
|
6S38
 
 | | Ligand binding domain of the P. putida receptor PcaY_PP in complex with quinate | | Descriptor: | (1S,3R,4S,5R)-1,3,4,5-tetrahydroxycyclohexanecarboxylic acid, Aromatic acid chemoreceptor | | Authors: | Gavira, J.A, Mantilla, M.A, Fernandez, M, Krell, T. | | Deposit date: | 2019-06-24 | | Release date: | 2020-10-21 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | The structural basis for signal promiscuity in a bacterial chemoreceptor.
Febs J., 288, 2021
|
|
6S37
 
 | | Ligand binding domain of the P. putida receptor PcaY_PP in complex with salicylic acid | | Descriptor: | 2-HYDROXYBENZOIC ACID, ACETATE ION, Aromatic acid chemoreceptor | | Authors: | Gavira, J.A, Mantilla, M.A, Fernandez, M, Krell, T. | | Deposit date: | 2019-06-24 | | Release date: | 2020-10-21 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The structural basis for signal promiscuity in a bacterial chemoreceptor.
Febs J., 288, 2021
|
|
6S18
 
 | | Ligand binding domain of the P. putida receptor PcaY_PP in complex with glycerol | | Descriptor: | Aromatic acid chemoreceptor, CHLORIDE ION, GLYCEROL | | Authors: | Gavira, J.A, Mantilla, M.A, Fernandez, M, Krell, T. | | Deposit date: | 2019-06-18 | | Release date: | 2020-10-21 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The structural basis for signal promiscuity in a bacterial chemoreceptor.
Febs J., 288, 2021
|
|
6S1A
 
 | | Ligand binding domain of the P. putida receptor PcaY_PP | | Descriptor: | Aromatic acid chemoreceptor, SULFATE ION | | Authors: | Gavira, J.A, Matilla, M.A, Fernandez, M, Krell, T. | | Deposit date: | 2019-06-18 | | Release date: | 2020-10-21 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.112 Å) | | Cite: | The structural basis for signal promiscuity in a bacterial chemoreceptor.
Febs J., 288, 2021
|
|
6S3B
 
 | | Ligand binding domain of the P. putida receptor PcaY_PP in complex with benzoate | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Aromatic acid chemoreceptor, ... | | Authors: | Gavira, J.A, Mantilla, M.A, Fernandez, M, Krell, T. | | Deposit date: | 2019-06-24 | | Release date: | 2020-10-21 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The structural basis for signal promiscuity in a bacterial chemoreceptor.
Febs J., 288, 2021
|
|
4BA7
 
 | | Crystal Structure of Ancestral Thioredoxin Relative to Last Bacteria Common Ancestor (LBCA) from the Precambrian Period | | Descriptor: | 1,2-ETHANEDIOL, LBCA THIOREDOXIN | | Authors: | Gavira, J.A, Ingles-Prieto, A, Ibarra-Molero, B, Sanchez-Ruiz, J.M. | | Deposit date: | 2012-09-12 | | Release date: | 2013-08-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Conservation of Protein Structure Over Four Billion Years
Structure, 21, 2013
|
|
6YMZ
 
 | | Structure of the CheB methylsterase from P. atrosepticum SCRI1043 | | Descriptor: | ACETATE ION, GLYCEROL, Protein-glutamate methylesterase/protein-glutamine glutaminase, ... | | Authors: | Gavira, J.A, Krell, T, Velando-Soriano, F, Matilla, M.A. | | Deposit date: | 2020-04-10 | | Release date: | 2021-02-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Evidence for Pentapeptide-Dependent and Independent CheB Methylesterases.
Int J Mol Sci, 21, 2020
|
|
