7UAV
 
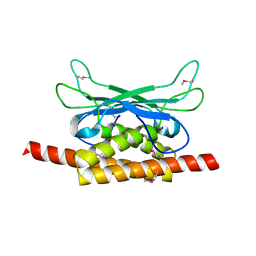 | | Structure of Clostridium botulinum prophage Tad1 in apo state | | Descriptor: | ABC transporter ATPase | | Authors: | Lu, A, Leavitt, A, Yirmiya, E, Amitai, G, Garb, J, Morehouse, B.R, Hobbs, S.J, Sorek, R, Kranzusch, P.J. | | Deposit date: | 2022-03-14 | | Release date: | 2022-10-05 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Viruses inhibit TIR gcADPR signalling to overcome bacterial defence.
Nature, 611, 2022
|
|
7UAW
 
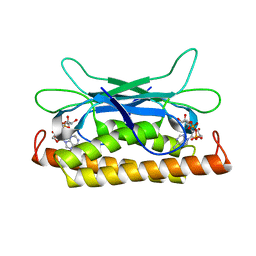 | | Structure of Clostridium botulinum prophage Tad1 in complex with 1''-2' gcADPR | | Descriptor: | (1S,3R,4R,6R,9S,11R,14R,15S,16R,18R)-4-(6-amino-9H-purin-9-yl)-9,11,15,16,18-pentahydroxy-2,5,8,10,12,17-hexaoxa-9lambda~5~,11lambda~5~-diphosphatricyclo[12.2.1.1~3,6~]octadecane-9,11-dione, ABC transporter ATPase | | Authors: | Lu, A, Leavitt, A, Yirmiya, E, Amitai, G, Garb, J, Morehouse, B.R, Hobbs, S.J, Sorek, R, Kranzusch, P.J. | | Deposit date: | 2022-03-14 | | Release date: | 2022-10-05 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Viruses inhibit TIR gcADPR signalling to overcome bacterial defence.
Nature, 611, 2022
|
|
9CS7
 
 | | Structure of Azospirillum bacterial CARD crystal form 1 | | Descriptor: | Serine protease | | Authors: | Wein, T, Millman, A, Lange, K, Yirmiya, E, Hadary, R, Garb, J, Melamed, S, Steinruecke, F, HIll, A.B, Kranzusch, P.J, Sorek, R. | | Deposit date: | 2024-07-23 | | Release date: | 2025-01-29 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | CARD domains mediate anti-phage defence in bacterial gasdermin systems.
Nature, 639, 2025
|
|
9CS8
 
 | | Structure of Azospirillum bacterial CARD crystal form 2 | | Descriptor: | Serine protease | | Authors: | Wein, T, Millman, A, Lange, K, Yirmiya, E, Hadary, R, Garb, J, Melamed, S, Steinruecke, F, Hill, A.B, Kranzusch, P.J, Sorek, R. | | Deposit date: | 2024-07-23 | | Release date: | 2025-01-29 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | CARD domains mediate anti-phage defence in bacterial gasdermin systems.
Nature, 639, 2025
|
|
8SMF
 
 | | Structure of SPO1 phage Tad2 in complex with 1''-3' gcADPR | | Descriptor: | (2R,3R,3aS,5S,6R,7S,8R,11R,13S,15aR)-2-(6-amino-9H-purin-9-yl)-3,6,7,11,13-pentahydroxyoctahydro-2H,5H,11H,13H-5,8-epoxy-11lambda~5~,13lambda~5~-furo[2,3-g][1,3,5,9,2,4]tetraoxadiphosphacyclotetradecine-11,13-dione, Gp34.65, MAGNESIUM ION | | Authors: | Lu, A, Yirmiya, E, Leavitt, A, Avraham, C, Osterman, I, Garb, J, Antine, S.P, Mooney, S.E, Hobbs, S.J, Amitai, G, Sorek, R, Kranzusch, P.J. | | Deposit date: | 2023-04-26 | | Release date: | 2023-11-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Phages overcome bacterial immunity via diverse anti-defence proteins.
Nature, 625, 2024
|
|
8SMG
 
 | | Structure of SPO1 phage Tad2 in complex with 1''-2' gcADPR | | Descriptor: | (1S,3R,4R,6R,9S,11R,14R,15S,16R,18R)-4-(6-amino-9H-purin-9-yl)-9,11,15,16,18-pentahydroxy-2,5,8,10,12,17-hexaoxa-9lambda~5~,11lambda~5~-diphosphatricyclo[12.2.1.1~3,6~]octadecane-9,11-dione, Gp34.65 | | Authors: | Lu, A, Yirmiya, E, Leavitt, A, Avraham, C, Osterman, I, Garb, J, Antine, S.P, Mooney, S.E, Hobbs, S.J, Amitai, G, Sorek, R, Kranzusch, P.J. | | Deposit date: | 2023-04-26 | | Release date: | 2023-11-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Phages overcome bacterial immunity via diverse anti-defence proteins.
Nature, 625, 2024
|
|
8SMD
 
 | | Structure of Clostridium botulinum prophage Tad1 in complex with 1''-3' gcADPR | | Descriptor: | (2R,3R,3aS,5S,6R,7S,8R,11R,13S,15aR)-2-(6-amino-9H-purin-9-yl)-3,6,7,11,13-pentahydroxyoctahydro-2H,5H,11H,13H-5,8-epoxy-11lambda~5~,13lambda~5~-furo[2,3-g][1,3,5,9,2,4]tetraoxadiphosphacyclotetradecine-11,13-dione, ABC transporter ATPase | | Authors: | Lu, A, Yirmiya, E, Leavitt, A, Avraham, C, Osterman, I, Garb, J, Antine, S.P, Mooney, S.E, Hobbs, S.J, Amitai, G, Sorek, R, Kranzusch, P.J. | | Deposit date: | 2023-04-26 | | Release date: | 2023-11-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Phages overcome bacterial immunity via diverse anti-defence proteins.
Nature, 625, 2024
|
|
8SME
 
 | | Structure of SPO1 phage Tad2 in apo state | | Descriptor: | Gp34.65 | | Authors: | Lu, A, Yirmiya, E, Leavitt, A, Avraham, C, Osterman, I, Garb, J, Antine, S.P, Mooney, S.E, Hobbs, S.J, Amitai, G, Sorek, R, Kranzusch, P.J. | | Deposit date: | 2023-04-26 | | Release date: | 2023-11-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Phages overcome bacterial immunity via diverse anti-defence proteins.
Nature, 625, 2024
|
|
1OKQ
 
 | | LAMININ ALPHA 2 CHAIN LG4-5 DOMAIN PAIR, CA1 SITE MUTANT | | Descriptor: | CALCIUM ION, LAMININ ALPHA 2 CHAIN | | Authors: | Wizemann, H, Garbe, J.H.O, Friedrich, M.V.K, Timpl, R, Sasaki, T, Hohenester, E. | | Deposit date: | 2003-07-28 | | Release date: | 2003-09-11 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Distinct Requirements for Heparin and Alpha-Dystroglycan Binding Revealed by Structure-Based Mutagenesis of the Laminin Alpha2 Lg4-Lg5 Domain Pair
J.Mol.Biol., 332, 2003
|
|
8U5O
 
 | | The structure of the catalytic domain of NanI sialdase in complex with Neu5Gc | | Descriptor: | CALCIUM ION, Exo-alpha-sialidase, N-glycolyl-alpha-neuraminic acid, ... | | Authors: | Medley, B.J, Low, K.E, Garber, J.M, Gray, T.E, Liu, L, Klassen, L, Fordwour, O.B, Inglis, G.D, Boons, G.J, Zandberg, W.F, Abbott, W.D, Boraston, A.B. | | Deposit date: | 2023-09-12 | | Release date: | 2024-09-04 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | A "terminal" case of glycan catabolism: Structural and enzymatic characterization of the sialidases of Clostridium perfringens.
J.Biol.Chem., 300, 2024
|
|
9C20
 
 | | The Sialidase NanJ in complex with Neu5,9Ac | | Descriptor: | 1,2-ETHANEDIOL, 9-O-acetyl-5-acetamido-3,5-dideoxy-D-glycero-alpha-D-galacto-non-2-ulopyranosonic acid, exo-alpha-sialidase | | Authors: | Medley, B.J, Low, K.E, Garber, J.M, Gray, T.E, Leeann, L.L, Fordwour, O.B, Inglis, G.D, Boons, G.J, Zandberg, W.F, Abbott, W, Boraston, A. | | Deposit date: | 2024-05-30 | | Release date: | 2024-09-04 | | Last modified: | 2025-03-19 | | Method: | X-RAY DIFFRACTION (2.699 Å) | | Cite: | A "terminal" case of glycan catabolism: Structural and enzymatic characterization of the sialidases of Clostridium perfringens.
J.Biol.Chem., 300, 2024
|
|
