7X2U
 
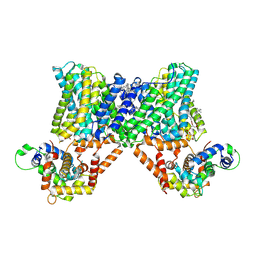 | | Structure of a human NHE3-CHP1 complex in the autoinhibited state | | Descriptor: | (1S)-2-{[{[(2R)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL STEARATE, Calcineurin B homologous protein 1, Sodium/hydrogen exchanger 3, ... | | Authors: | Dong, Y, Li, H, Gao, Y, Zhang, X.C, Zhao, Y. | | Deposit date: | 2022-02-26 | | Release date: | 2022-04-20 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structure of a human NHE3-CHP1 complex in the autoinhibited state
Sci Adv, 2022
|
|
7XDK
 
 | | Cryo-EM structure of SARS-CoV-2 Delta Spike protein in complex with BA7054 and BA7125 fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BA7054 fab, ... | | Authors: | Liu, Z, Lui, S, Gao, Y. | | Deposit date: | 2022-03-27 | | Release date: | 2023-03-01 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Biparatopic antibody BA7208/7125 effectively neutralizes SARS-CoV-2 variants including Omicron BA.1-BA.5.
Cell Discov, 9, 2023
|
|
8K90
 
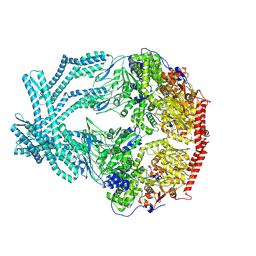 | | CryoEM structure of LonC protease open pentamer in presence of AGS | | Descriptor: | Monothiophosphate, endopeptidase La | | Authors: | Li, M, Hsieh, K, Liu, H, Zhang, S, Gao, Y, Gong, Q, Zhang, K, Chang, C, Li, S. | | Deposit date: | 2023-07-31 | | Release date: | 2024-08-07 | | Method: | ELECTRON MICROSCOPY (3.43 Å) | | Cite: | Bifurcated assembly pathway and dual function of a Lon-like protease revealed by cryo-EM Analysis
Fundam Res, 2024
|
|
8K8V
 
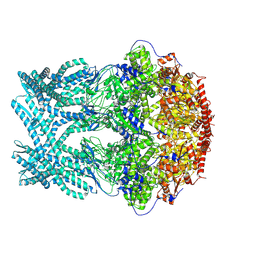 | | CryoEM structure of LonC protease hepatmer, apo state | | Descriptor: | PHOSPHATE ION, endopeptidase La | | Authors: | Li, M, Hsieh, K, Liu, H, Zhang, S, Gao, Y, Gong, Q, Zhang, K, Chang, C, Li, S. | | Deposit date: | 2023-07-31 | | Release date: | 2024-08-07 | | Method: | ELECTRON MICROSCOPY (2.75 Å) | | Cite: | Bifurcated assembly pathway and dual function of a Lon-like protease revealed by cryo-EM Analysis
Fundam Res, 2024
|
|
8K8Z
 
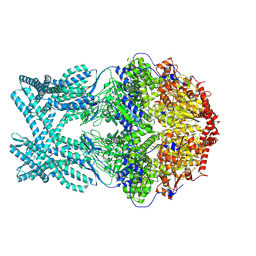 | | CryoEM structure of LonC protease hexamer in presence of AGS | | Descriptor: | Monothiophosphate, endopeptidase La | | Authors: | Li, M, Hsieh, K, Liu, H, Zhang, S, Gao, Y, Gong, Q, Zhang, K, Chang, C, Li, S. | | Deposit date: | 2023-07-31 | | Release date: | 2024-08-07 | | Method: | ELECTRON MICROSCOPY (2.83 Å) | | Cite: | Bifurcated assembly pathway and dual function of a Lon-like protease revealed by cryo-EM Analysis
Fundam Res, 2024
|
|
8K92
 
 | | CryoEM structure of LonC S582A hexamer with Lysozyme | | Descriptor: | Endopeptidase La, Monothiophosphate | | Authors: | Li, M, Hsieh, K, Liu, H, Zhang, S, Gao, Y, Gong, Q, Zhang, K, Chang, C, Li, S. | | Deposit date: | 2023-07-31 | | Release date: | 2024-08-07 | | Method: | ELECTRON MICROSCOPY (3.31 Å) | | Cite: | Bifurcated assembly pathway and dual function of a Lon-like protease revealed by cryo-EM Analysis
Fundam Res, 2024
|
|
8K95
 
 | | CryoEM structure of LonC protease open Hexamer, AGS | | Descriptor: | Monothiophosphate, endopeptidase La | | Authors: | Li, M, Hsieh, K, Liu, H, Zhang, S, Gao, Y, Gong, Q, Zhang, K, Chang, C, Li, S. | | Deposit date: | 2023-07-31 | | Release date: | 2024-08-07 | | Method: | ELECTRON MICROSCOPY (3.05 Å) | | Cite: | Bifurcated assembly pathway and dual function of a Lon-like protease revealed by cryo-EM Analysis
Fundam Res, 2024
|
|
8K91
 
 | | CryoEM structure of LonC S582A hepatmer with Lysozyme | | Descriptor: | Endopeptidase La, Monothiophosphate | | Authors: | Li, M, Hsieh, K, Liu, H, Zhang, S, Gao, Y, Gong, Q, Zhang, K, Chang, C, Li, S. | | Deposit date: | 2023-07-31 | | Release date: | 2024-08-07 | | Method: | ELECTRON MICROSCOPY (3.03 Å) | | Cite: | Bifurcated assembly pathway and dual function of a Lon-like protease revealed by cryo-EM Analysis
Fundam Res, 2024
|
|
8K93
 
 | | CryoEM structure of LonC protease S582A open hexamer with lysozyme | | Descriptor: | Endopeptidase La, Monothiophosphate | | Authors: | Li, M, Hsieh, K, Liu, H, Zhang, S, Gao, Y, Gong, Q, Zhang, K, Chang, C, Li, S. | | Deposit date: | 2023-07-31 | | Release date: | 2024-08-07 | | Method: | ELECTRON MICROSCOPY (3.23 Å) | | Cite: | Bifurcated assembly pathway and dual function of a Lon-like protease revealed by cryo-EM Analysis
Fundam Res, 2024
|
|
8K8Y
 
 | | CryoEM structure of LonC heptamer in presence of AGS | | Descriptor: | Monothiophosphate, endopeptidase La | | Authors: | Li, M, Hsieh, K, Liu, H, Zhang, S, Gao, Y, Gong, Q, Zhang, K, Chang, C, Li, S. | | Deposit date: | 2023-07-31 | | Release date: | 2024-08-07 | | Method: | ELECTRON MICROSCOPY (2.54 Å) | | Cite: | Bifurcated assembly pathway and dual function of a Lon-like protease revealed by cryo-EM Analysis
Fundam Res, 2024
|
|
8K8W
 
 | | CryoEM structure of LonC protease open hexamer, apo state | | Descriptor: | endopeptidase La | | Authors: | Li, M, Hsieh, K, Liu, H, Zhang, S, Gao, Y, Gong, Q, Zhang, K, Chang, C, Li, S. | | Deposit date: | 2023-07-31 | | Release date: | 2024-08-07 | | Method: | ELECTRON MICROSCOPY (3.46 Å) | | Cite: | Bifurcated assembly pathway and dual function of a Lon-like protease revealed by cryo-EM Analysis
Fundam Res, 2024
|
|
8K96
 
 | | CryoEM structure of LonC protease hepatmer with Bortezomib | | Descriptor: | N-[(1R)-1-(DIHYDROXYBORYL)-3-METHYLBUTYL]-N-(PYRAZIN-2-YLCARBONYL)-L-PHENYLALANINAMIDE, PHOSPHATE ION, endopeptidase La | | Authors: | Li, M, Hsieh, K, Liu, H, Zhang, S, Gao, Y, Gong, Q, Zhang, K, Chang, C, Li, S. | | Deposit date: | 2023-07-31 | | Release date: | 2024-08-07 | | Method: | ELECTRON MICROSCOPY (2.89 Å) | | Cite: | Bifurcated assembly pathway and dual function of a Lon-like protease revealed by cryo-EM Analysis
Fundam Res, 2024
|
|
8K97
 
 | | CryoEM structure of LonC protease hexamer with Bortezomib | | Descriptor: | N-[(1R)-1-(DIHYDROXYBORYL)-3-METHYLBUTYL]-N-(PYRAZIN-2-YLCARBONYL)-L-PHENYLALANINAMIDE, PHOSPHATE ION, endopeptidase La | | Authors: | Li, M, Hsieh, K, Liu, H, Zhang, S, Gao, Y, Gong, Q, Zhang, K, Chang, C, Li, S. | | Deposit date: | 2023-07-31 | | Release date: | 2024-08-07 | | Method: | ELECTRON MICROSCOPY (2.73 Å) | | Cite: | Bifurcated assembly pathway and dual function of a Lon-like protease revealed by cryo-EM Analysis
Fundam Res, 2024
|
|
8K8X
 
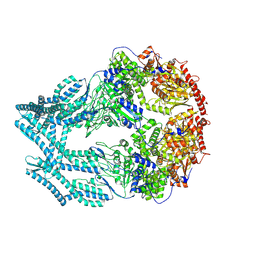 | | CryoEM of LonC open pentamer, apo state | | Descriptor: | endopeptidase La | | Authors: | Li, M, Hsieh, K, Liu, H, Zhang, S, Gao, Y, Gong, Q, Zhang, K, Chang, C, Li, S. | | Deposit date: | 2023-07-31 | | Release date: | 2024-08-07 | | Method: | ELECTRON MICROSCOPY (3.53 Å) | | Cite: | Bifurcated assembly pathway and dual function of a Lon-like protease revealed by cryo-EM Analysis
Fundam Res, 2024
|
|
8K94
 
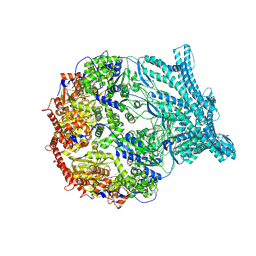 | | CryoEM structure of LonC protease S582A open pentamer with lysozyme | | Descriptor: | Endopeptidase La, Monothiophosphate | | Authors: | Li, M, Hsieh, K, Liu, H, Zhang, S, Gao, Y, Gong, Q, Zhang, K, Chang, C, Li, S. | | Deposit date: | 2023-07-31 | | Release date: | 2024-08-07 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Bifurcated assembly pathway and dual function of a Lon-like protease revealed by cryo-EM Analysis
Fundam Res, 2024
|
|
7WS1
 
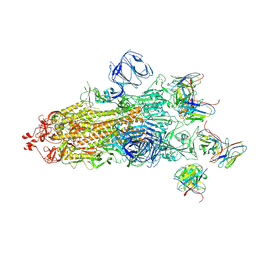 | | Structures of Omicron Spike complexes illuminate broad-spectrum neutralizing antibody development | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 510A5 heavy chain, ... | | Authors: | Guo, H, Gao, Y, Ji, X, Yang, H. | | Deposit date: | 2022-01-28 | | Release date: | 2022-06-01 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structures of Omicron spike complexes and implications for neutralizing antibody development.
Cell Rep, 39, 2022
|
|
7WS2
 
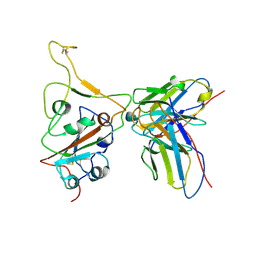 | | Structures of Omicron Spike complexes illuminate broad-spectrum neutralizing antibody development | | Descriptor: | 510A5 heavy chain, 510A5 light chain, Spike protein S1 | | Authors: | Guo, H, Gao, Y, Ji, X, Yang, H. | | Deposit date: | 2022-01-28 | | Release date: | 2022-06-01 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structures of Omicron spike complexes and implications for neutralizing antibody development.
Cell Rep, 39, 2022
|
|
7WS8
 
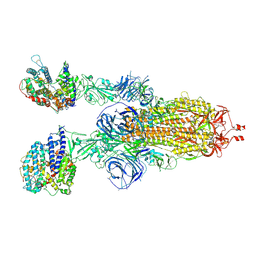 | | Structures of Omicron Spike complexes illuminate broad-spectrum neutralizing antibody development | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, ... | | Authors: | Guo, H, Gao, Y, Ji, X, Yang, H. | | Deposit date: | 2022-01-28 | | Release date: | 2022-06-01 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structures of Omicron spike complexes and implications for neutralizing antibody development.
Cell Rep, 39, 2022
|
|
7WS0
 
 | | Structures of Omicron Spike complexes illuminate broad-spectrum neutralizing antibody development | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 510A5 heavy chain, ... | | Authors: | Guo, H, Gao, Y, Ji, X, Yang, H. | | Deposit date: | 2022-01-28 | | Release date: | 2022-06-01 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structures of Omicron spike complexes and implications for neutralizing antibody development.
Cell Rep, 39, 2022
|
|
7WS6
 
 | | Structures of Omicron Spike complexes illuminate broad-spectrum neutralizing antibody development | | Descriptor: | 510A5 heavy chain, 510A5 light chain, Spike protein S1 | | Authors: | Guo, H, Gao, Y, Ji, X, Yang, H. | | Deposit date: | 2022-01-28 | | Release date: | 2022-06-01 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structures of Omicron spike complexes and implications for neutralizing antibody development.
Cell Rep, 39, 2022
|
|
7WSA
 
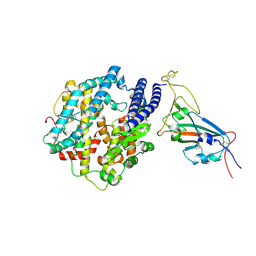 | | Structures of Omicron Spike complexes illuminate broad-spectrum neutralizing antibody development | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, Spike protein S1 | | Authors: | Guo, H, Gao, Y, Ji, X, Yang, H. | | Deposit date: | 2022-01-28 | | Release date: | 2022-06-01 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structures of Omicron spike complexes and implications for neutralizing antibody development.
Cell Rep, 39, 2022
|
|
7WS7
 
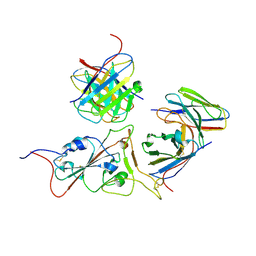 | | Structures of Omicron Spike complexes illuminate broad-spectrum neutralizing antibody development | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 510A5 heavy chain, 510A5 light chain, ... | | Authors: | Guo, H, Gao, Y, Ji, X, Yang, H. | | Deposit date: | 2022-01-28 | | Release date: | 2022-06-01 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structures of Omicron spike complexes and implications for neutralizing antibody development.
Cell Rep, 39, 2022
|
|
7WS9
 
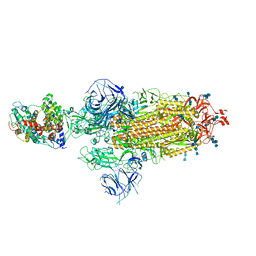 | | Structures of Omicron Spike complexes illuminate broad-spectrum neutralizing antibody development | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, ... | | Authors: | Guo, H, Gao, Y, Ji, X, Yang, H. | | Deposit date: | 2022-01-28 | | Release date: | 2022-06-01 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structures of Omicron spike complexes and implications for neutralizing antibody development.
Cell Rep, 39, 2022
|
|
7WS3
 
 | | Structures of Omicron Spike complexes illuminate broad-spectrum neutralizing antibody development | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 510A5 heavy chain, ... | | Authors: | Guo, H, Gao, Y, Ji, X, Yang, H. | | Deposit date: | 2022-01-28 | | Release date: | 2022-06-01 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structures of Omicron spike complexes and implications for neutralizing antibody development.
Cell Rep, 39, 2022
|
|
7WS4
 
 | | Ultrapotent SARS-CoV-2 neutralizing antibodies with protective efficacy against newly emerged mutational variants | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 510A5 heavy chain, ... | | Authors: | Guo, H, Gao, Y, Ji, X, Yang, H. | | Deposit date: | 2022-01-28 | | Release date: | 2022-06-01 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structures of Omicron spike complexes and implications for neutralizing antibody development.
Cell Rep, 39, 2022
|
|
