4K63
 
 | | Structure of an avian influenza H5 hemagglutinin from the influenza virus complexed with avian receptor analog LSTa | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin, N-acetyl-alpha-neuraminic acid-(2-3)-beta-D-galactopyranose, ... | | Authors: | Zhang, W, Shi, Y, Lu, X, Shu, Y, Qi, J, Gao, G.F. | | Deposit date: | 2013-04-15 | | Release date: | 2013-05-22 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | An airborne transmissible avian influenza H5 hemagglutinin seen at the atomic level.
Science, 340, 2013
|
|
7W8S
 
 | | Structure of SARS-CoV-2 spike receptor-binding domain Y453F mutation complexed with American mink ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, Spike protein S1, ... | | Authors: | Su, C, Qi, J.X, Gao, G.F. | | Deposit date: | 2021-12-08 | | Release date: | 2022-08-17 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.85 Å) | | Cite: | Molecular Basis of Mink ACE2 Binding to SARS-CoV-2 and Its Mink-Derived Variants.
J.Virol., 96, 2022
|
|
7WA3
 
 | | Structure of American mink ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme, ... | | Authors: | Su, C, Qi, J.X, Gao, G.F. | | Deposit date: | 2021-12-11 | | Release date: | 2022-08-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | The molecular basis of SARS-CoV-2 variants binding to mink ACE2
To Be Published
|
|
7WA1
 
 | |
4K65
 
 | | Structure of an airborne transmissible avian influenza H5 hemagglutinin mutant from the influenza virus A/Indonesia/5/2005 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin | | Authors: | Zhang, W, Shi, Y, Lu, X, Shu, Y, Qi, J, Gao, G.F. | | Deposit date: | 2013-04-15 | | Release date: | 2013-05-22 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | An airborne transmissible avian influenza H5 hemagglutinin seen at the atomic level.
Science, 340, 2013
|
|
4KDQ
 
 | | Crystal structure of the hemagglutinin of A/Xinjiang/1/2006 virus | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin | | Authors: | Lu, X, Shi, Y, Zhang, W, Zhang, Y, Qi, J, Gao, G.F. | | Deposit date: | 2013-04-25 | | Release date: | 2013-07-24 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.604 Å) | | Cite: | Structure and receptor-binding properties of an airborne transmissible avian influenza A virus hemagglutinin H5 (VN1203mut).
Protein Cell, 4, 2013
|
|
4LLA
 
 | | Crystal structure of D3D4 domain of the LILRB2 molecule | | Descriptor: | Leukocyte immunoglobulin-like receptor subfamily B member 2 | | Authors: | Nam, G, Shi, Y, Ryu, M, Wang, Q, Song, H, Liu, J, Yan, J, Qi, J, Gao, G.F. | | Deposit date: | 2013-07-09 | | Release date: | 2013-09-11 | | Last modified: | 2013-11-06 | | Method: | X-RAY DIFFRACTION (2.502 Å) | | Cite: | Crystal structures of the two membrane-proximal Ig-like domains (D3D4) of LILRB1/B2: alternative models for their involvement in peptide-HLA binding
Protein Cell, 4, 2013
|
|
4LNR
 
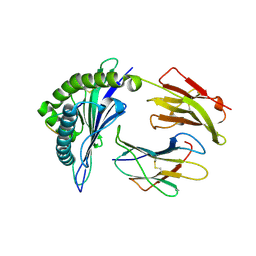 | | The structure of HLA-B*35:01 in complex with the peptide (RPQVPLRPMTY) | | Descriptor: | Beta-2-microglobulin, HLA class I histocompatibility antigen, B-35 alpha chain, ... | | Authors: | Cheng, H, Shi, Y, Qi, J, Gao, G.F. | | Deposit date: | 2013-07-12 | | Release date: | 2014-07-23 | | Last modified: | 2019-12-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Peptide-dependent conformational fluctuation determines the stability of the human leukocyte antigen class I complex.
J.Biol.Chem., 289, 2014
|
|
4LL9
 
 | | Crystal structure of D3D4 domain of the LILRB1 molecule | | Descriptor: | IODIDE ION, Leukocyte immunoglobulin-like receptor subfamily B member 1 | | Authors: | Nam, G, Shi, Y, Ryu, M, Wang, Q, Song, H, Liu, J, Yan, J, Qi, J, Gao, G.F. | | Deposit date: | 2013-07-09 | | Release date: | 2013-09-11 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.686 Å) | | Cite: | Crystal structures of the two membrane-proximal Ig-like domains (D3D4) of LILRB1/B2: alternative models for their involvement in peptide-HLA binding
Protein Cell, 4, 2013
|
|
4K62
 
 | | Structure of an avian influenza H5 hemagglutinin from the influenza virus A/Indonesia/5/2005 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin | | Authors: | Zhang, W, Shi, Y, Lu, X, Shu, Y, Qi, J, Gao, G.F. | | Deposit date: | 2013-04-15 | | Release date: | 2013-05-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.502 Å) | | Cite: | An airborne transmissible avian influenza H5 hemagglutinin seen at the atomic level.
Science, 340, 2013
|
|
4K66
 
 | | Structure of an airborne transmissible avian influenza H5 hemagglutinin mutant from the influenza virus A/Indonesia/5/2005 complexed with avian receptor analog LSTa | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin, N-acetyl-alpha-neuraminic acid-(2-3)-beta-D-galactopyranose | | Authors: | Zhang, W, Shi, Y, Lu, X, Shu, Y, Qi, J, Gao, G.F. | | Deposit date: | 2013-04-15 | | Release date: | 2013-05-22 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.005 Å) | | Cite: | An airborne transmissible avian influenza H5 hemagglutinin seen at the atomic level.
Science, 340, 2013
|
|
4KDN
 
 | | Crystal structure of the hemagglutinin of ferret-transmissible H5N1 virus in complex with avian receptor analog LSTa | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin, ... | | Authors: | Lu, X, Shi, Y, Zhang, W, Zhang, Y, Qi, J, Gao, G.F. | | Deposit date: | 2013-04-25 | | Release date: | 2013-07-24 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.483 Å) | | Cite: | Structure and receptor-binding properties of an airborne transmissible avian influenza A virus hemagglutinin H5 (VN1203mut).
Protein Cell, 4, 2013
|
|
4KDM
 
 | | Crystal structure of the hemagglutinin of ferret-transmissible H5N1 virus | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin | | Authors: | Lu, X, Shi, Y, Zhang, W, Zhang, Y, Qi, J, Gao, G.F. | | Deposit date: | 2013-04-25 | | Release date: | 2013-07-24 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.496 Å) | | Cite: | Structure and receptor-binding properties of an airborne transmissible avian influenza A virus hemagglutinin H5 (VN1203mut).
Protein Cell, 4, 2013
|
|
4I48
 
 | | Structure of HLA-A68 complexed with an HIV Env derived peptide | | Descriptor: | 9-mer peptide from Envelope glycoprotein gp160, Beta-2-microglobulin, HLA class I histocompatibility antigen, ... | | Authors: | Niu, L, Cheng, H, Zhang, S, Tan, S, Zhang, Y, Qi, J, Liu, J, Gao, G.F. | | Deposit date: | 2012-11-27 | | Release date: | 2013-10-02 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.799 Å) | | Cite: | Structural basis for the differential classification of HLA-A*6802 and HLA-A*6801 into the A2 and A3 supertypes
Mol.Immunol., 55, 2013
|
|
4I00
 
 | | Crystal structure of influenza A neuraminidase N3-H274Y complexed with zanamivir | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Neuraminidase, ... | | Authors: | Li, Q, Qi, J, Vavricka, C.J, Gao, G.F. | | Deposit date: | 2012-11-16 | | Release date: | 2013-11-13 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Functional and structural analysis of influenza virus neuraminidase N3 offers further insight into the mechanisms of oseltamivir resistance.
J.Virol., 87, 2013
|
|
4K64
 
 | | Structure of an avian influenza H5 hemagglutinin from the influenza virus complexed with human receptor analog LSTc | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin, N-acetyl-alpha-neuraminic acid-(2-6)-beta-D-galactopyranose | | Authors: | Zhang, W, Shi, Y, Lu, X, Shu, Y, Qi, J, Gao, G.F. | | Deposit date: | 2013-04-15 | | Release date: | 2013-05-22 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.604 Å) | | Cite: | An airborne transmissible avian influenza H5 hemagglutinin seen at the atomic level.
Science, 340, 2013
|
|
8WP8
 
 | | Cryo-EM structure of SARS-CoV-2 Omicron BA.2.86 RBD in complex with human ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, ... | | Authors: | Li, L.J, Gu, Y.H, Qi, J.X, Gao, G.F. | | Deposit date: | 2023-10-09 | | Release date: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (2.89 Å) | | Cite: | Cryo-EM structure of SARS-CoV-2 Omicron BA.2.86 RBD in complex with human ACE2
To Be Published
|
|
8XLV
 
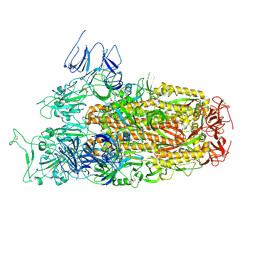 | | Cryo-EM structure of SARS-CoV-2 Omicron BA.2.86 spike protein(6P), 1-RBD-up state | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Li, L.J, Gu, Y.H, Shi, K.Y, Qi, J.X, Gao, G.F. | | Deposit date: | 2023-12-26 | | Release date: | 2024-07-03 | | Last modified: | 2024-08-21 | | Method: | ELECTRON MICROSCOPY (3.07 Å) | | Cite: | Spike structures, receptor binding, and immune escape of recently circulating SARS-CoV-2 Omicron BA.2.86, JN.1, EG.5, EG.5.1, and HV.1 sub-variants.
Structure, 32, 2024
|
|
8WOY
 
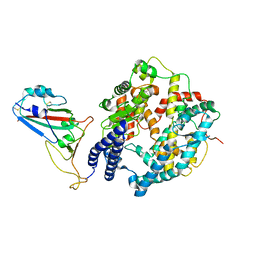 | | Cryo-EM structure of SARS-CoV-2 Omicron BA.4/5 RBD in complex with rabbit ACE2 (local refinement) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme, Spike protein S1, ... | | Authors: | Li, L.J, Shi, K.Y, Yu, G.H, Gao, G.F. | | Deposit date: | 2023-10-08 | | Release date: | 2023-12-13 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (3.14 Å) | | Cite: | Structural basis of increased binding affinities of spikes from SARS-CoV-2 Omicron variants to rabbit and hare ACE2s reveals the expanding host tendency.
Mbio, 15, 2024
|
|
8WOZ
 
 | | Cryo-EM structure of SARS-CoV RBD in complex with rabbit ACE2 | | Descriptor: | Angiotensin-converting enzyme, Spike protein S1, ZINC ION | | Authors: | Li, L.J, Shi, K.Y, Yu, G.H, Gao, G.F. | | Deposit date: | 2023-10-08 | | Release date: | 2023-12-13 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.25 Å) | | Cite: | Structural basis of increased binding affinities of spikes from SARS-CoV-2 Omicron variants to rabbit and hare ACE2s reveals the expanding host tendency.
Mbio, 15, 2024
|
|
8WOX
 
 | | Cryo-EM structure of SARS-CoV-2 prototype RBD in complex with rabbit ACE2 (local refinement) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme, Spike protein S1, ... | | Authors: | Li, L.J, Shi, K.Y, Yu, G.H, Gao, G.F. | | Deposit date: | 2023-10-08 | | Release date: | 2023-12-13 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (2.75 Å) | | Cite: | Structural basis of increased binding affinities of spikes from SARS-CoV-2 Omicron variants to rabbit and hare ACE2s reveals the expanding host tendency.
Mbio, 15, 2024
|
|
8XB8
 
 | | The structure of ASFV A137R | | Descriptor: | A137R | | Authors: | Li, C, Song, H, Gao, G.F. | | Deposit date: | 2023-12-06 | | Release date: | 2024-01-31 | | Last modified: | 2024-04-03 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | African swine fever virus A137R assembles into a dodecahedron cage.
J.Virol., 98, 2024
|
|
8XMT
 
 | | Cryo-EM structure of SARS-CoV-2 Omicron EG.5.1 spike protein(6P), RBD-closed state | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Li, L.J, Gu, Y.H, Shi, K.Y, Qi, J.X, Gao, G.F. | | Deposit date: | 2023-12-28 | | Release date: | 2024-07-03 | | Last modified: | 2024-08-21 | | Method: | ELECTRON MICROSCOPY (3.31 Å) | | Cite: | Spike structures, receptor binding, and immune escape of recently circulating SARS-CoV-2 Omicron BA.2.86, JN.1, EG.5, EG.5.1, and HV.1 sub-variants.
Structure, 32, 2024
|
|
8XN5
 
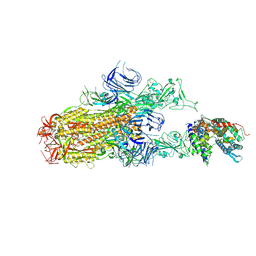 | | Cryo-EM structure of SARS-CoV-2 Omicron EG.5.1 spike protein(6P) in complex with human ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, Spike glycoprotein, ... | | Authors: | Li, L.J, Gu, Y.H, Shi, K.Y, Qi, J.X, Gao, G.F. | | Deposit date: | 2023-12-29 | | Release date: | 2024-07-03 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.87 Å) | | Cite: | Spike structures, receptor binding, and immune escape of recently circulating SARS-CoV-2 Omicron BA.2.86, JN.1, EG.5, EG.5.1, and HV.1 sub-variants.
Structure, 32, 2024
|
|
8XN3
 
 | | SARS-CoV-2 Omicron HV.1 RBD in complex with human ACE2 (local refinement from the spike protein) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, Spike protein S1, ... | | Authors: | Li, L.J, Gu, Y.H, Shi, K.Y, Qi, J.X, Gao, G.F. | | Deposit date: | 2023-12-28 | | Release date: | 2024-07-03 | | Last modified: | 2024-08-21 | | Method: | ELECTRON MICROSCOPY (2.64 Å) | | Cite: | Spike structures, receptor binding, and immune escape of recently circulating SARS-CoV-2 Omicron BA.2.86, JN.1, EG.5, EG.5.1, and HV.1 sub-variants.
Structure, 32, 2024
|
|
