6TEO
 
 | | Crystal structure of a yeast Snu114-Prp8 complex | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Pre-mRNA-splicing factor 8, ... | | Authors: | Ganichkin, O, Jia, J, Loll, B, Absmeier, E, Wahl, M.C. | | Deposit date: | 2019-11-12 | | Release date: | 2020-03-18 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | A Snu114-GTP-Prp8 module forms a relay station for efficient splicing in yeast.
Nucleic Acids Res., 48, 2020
|
|
2V9V
 
 | | Crystal Structure of Moorella thermoacetica SelB(377-511) | | Descriptor: | CHLORIDE ION, SELENOCYSTEINE-SPECIFIC ELONGATION FACTOR, SODIUM ION | | Authors: | Ganichkin, O, Wahl, M.C. | | Deposit date: | 2007-08-27 | | Release date: | 2007-09-11 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Conformational Switches in Winged-Helix Domains 1 and 2 of Bacterial Translation Elongation Factor Selb.
Acta Crystallogr.,Sect.D, 63, 2007
|
|
7S25
 
 | | ROCK1 IN COMPLEX WITH LIGAND G4998 | | Descriptor: | 2-[3-(methoxymethyl)phenyl]-N-[4-(1H-pyrazol-4-yl)phenyl]acetamide, CHLORIDE ION, Rho-associated protein kinase 1 | | Authors: | Ganichkin, O, Harris, S.F, Steinbacher, S. | | Deposit date: | 2021-09-03 | | Release date: | 2022-10-05 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.337 Å) | | Cite: | Chemical space docking enables large-scale structure-based virtual screening to discover ROCK1 kinase inhibitors.
Nat Commun, 13, 2022
|
|
7S26
 
 | | ROCK1 IN COMPLEX WITH LIGAND G5018 | | Descriptor: | 2-[methyl(phenyl)amino]-1-[4-(1H-pyrrolo[2,3-b]pyridin-3-yl)-3,6-dihydropyridin-1(2H)-yl]ethan-1-one, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Rho-associated protein kinase 1 | | Authors: | Ganichkin, O, Harris, S.F, Steinbacher, S. | | Deposit date: | 2021-09-03 | | Release date: | 2022-10-05 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.744 Å) | | Cite: | Chemical space docking enables large-scale structure-based virtual screening to discover ROCK1 kinase inhibitors.
Nat Commun, 13, 2022
|
|
6S9A
 
 | | Artificial GTPase-BSE dimer of human Dynamin1 | | Descriptor: | CHLORIDE ION, Dynamin-1,Dynamin-1, ZINC ION | | Authors: | Ganichkin, O.M, Vancraenenbroeck, R, Rosenblum, G, Hofmann, H, Daumke, O, Noel, J.K. | | Deposit date: | 2019-07-11 | | Release date: | 2020-08-26 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Quantification and demonstration of the collective constriction-by-ratchet mechanism in the dynamin molecular motor.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
3BCB
 
 | |
3BCA
 
 | |
3BC8
 
 | | Crystal structure of mouse selenocysteine synthase | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, O-phosphoseryl-tRNA(Sec) selenium transferase | | Authors: | Ganichkin, O.M, Wahl, M.C. | | Deposit date: | 2007-11-12 | | Release date: | 2007-12-18 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structure and catalytic mechanism of eukaryotic selenocysteine synthase.
J.Biol.Chem., 283, 2008
|
|
7OFV
 
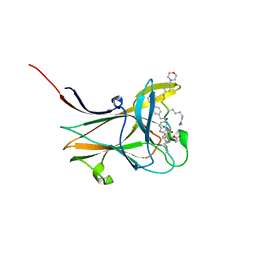 | | NMR-guided design of potent and selective EphA4 agonistic ligands | | Descriptor: | ACETATE ION, EphA4 agonist ligand, Ephrin type-A receptor 4 | | Authors: | Ganichkin, O.M, Craig, T.K, Baggio, C, Pellecchia, M. | | Deposit date: | 2021-05-05 | | Release date: | 2021-08-11 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | NMR-Guided Design of Potent and Selective EphA4 Agonistic Ligands.
J.Med.Chem., 64, 2021
|
|
9GWZ
 
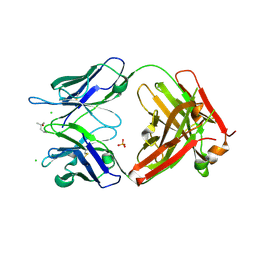 | | Crystal structure of 23ME-00610 Fab | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 23ME-00610 Fab (heavy), 23ME-00610 Fab (light), ... | | Authors: | Huang, Y.M, Ganichkin, O.M. | | Deposit date: | 2024-09-27 | | Release date: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | CD200R1 immune checkpoint blockade by the first-in-human anti-CD200R1 antibody 23ME-00610: molecular mechanism and engineering of a surrogate antibody.
Mabs, 16, 2024
|
|
9GWT
 
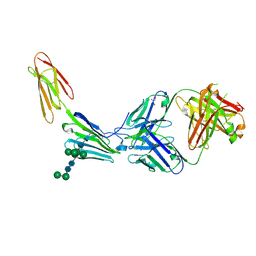 | | crystal structure of 23ME-00610 Fab in complex with human CD200R1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 23ME-00610 Fab (heavy), 23ME-00610 Fab (light), ... | | Authors: | Huang, Y.M, Ganichkin, O.M. | | Deposit date: | 2024-09-27 | | Release date: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.89 Å) | | Cite: | CD200R1 immune checkpoint blockade by the first-in-human anti-CD200R1 antibody 23ME-00610: molecular mechanism and engineering of a surrogate antibody.
Mabs, 16, 2024
|
|
8R7B
 
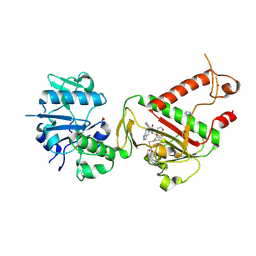 | | SARS-CoV-2 NSP14 in complex with SAH and TDI-015051 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Guanine-N7 methyltransferase nsp14, ... | | Authors: | Meyer, C, Garzia, A, Miller, M, Huggins, D.J, Myers, R.W, Liverton, N, Kargman, S, Nitsche, J, Ganichkin, O, Steinbacher, S, Meinke, P.T, Tuschl, T. | | Deposit date: | 2023-11-24 | | Release date: | 2024-10-23 | | Last modified: | 2025-02-05 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Small-molecule inhibition of SARS-CoV-2 NSP14 RNA cap methyltransferase.
Nature, 637, 2025
|
|
3RG5
 
 | | Crystal Structure of Mouse tRNA(Sec) | | Descriptor: | ACETATE ION, GLYCEROL, SULFATE ION, ... | | Authors: | Wahl, M.C, Ganichkin, O.M, Anedchenko, E.A. | | Deposit date: | 2011-04-07 | | Release date: | 2011-06-08 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure analysis reveals functional flexibility in the selenocysteine-specific tRNA from mouse.
Plos One, 6, 2011
|
|
