8CMS
 
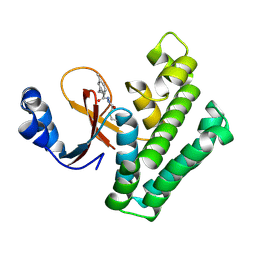 | | OTUB2 in covalent complex with LN5P45 | | Descriptor: | (1~{S},2~{S})-~{N}'-ethanoyl-2-(3-methylphenyl)cyclopropane-1-carbohydrazide, Ubiquitin thioesterase OTUB2 | | Authors: | Gan, J, de Vries, J. | | Deposit date: | 2023-02-21 | | Release date: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Cellular Validation of a Chemically Improved Inhibitor Identifies Monoubiquitination on OTUB2.
Acs Chem.Biol., 18, 2023
|
|
2G1J
 
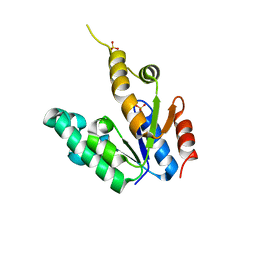 | | Crystal structure of Mycobacterium tuberculosis Shikimate Kinase at 2.0 angstrom resolution | | Descriptor: | SULFATE ION, Shikimate kinase | | Authors: | Gan, J, Gu, Y, Li, Y, Yan, H, Ji, X. | | Deposit date: | 2006-02-14 | | Release date: | 2006-07-18 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of Mycobacterium tuberculosis Shikimate Kinase in Complex with Shikimic Acid and an ATP Analogue.
Biochemistry, 45, 2006
|
|
8XIF
 
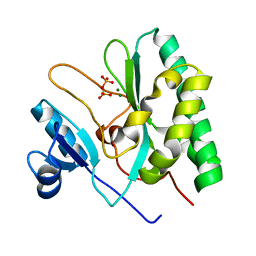 | | The crystal structure of the AEP domain of VACV D5 | | Descriptor: | MAGNESIUM ION, PYROPHOSPHATE, Uncoating factor OPG117 | | Authors: | Gan, J, Zhang, W. | | Deposit date: | 2023-12-19 | | Release date: | 2024-05-01 | | Last modified: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | Structural and functional insights into the helicase protein E5 of Mpox virus.
Cell Discov, 10, 2024
|
|
8XIG
 
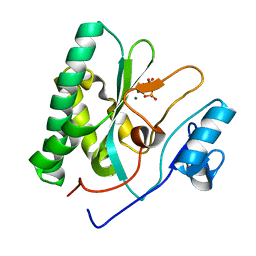 | | The crystal structure of the AEP domain of MPXV E5 | | Descriptor: | MAGNESIUM ION, PYROPHOSPHATE, Uncoating factor OPG117 | | Authors: | Gan, J, Zhang, W. | | Deposit date: | 2023-12-19 | | Release date: | 2024-05-01 | | Last modified: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural and functional insights into the helicase protein E5 of Mpox virus.
Cell Discov, 10, 2024
|
|
5JRE
 
 | | Crystal structure of NeC3PO in complex with ssDNA. | | Descriptor: | 9-METHYL-9H-PURIN-6-AMINE, ADENINE, NEQ131, ... | | Authors: | Gan, J, Zhang, J. | | Deposit date: | 2016-05-06 | | Release date: | 2016-09-28 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis for single-stranded RNA recognition and cleavage by C3PO
Nucleic Acids Res., 44, 2016
|
|
6R0J
 
 | |
2EZ6
 
 | | Crystal structure of Aquifex aeolicus RNase III (D44N) complexed with product of double-stranded RNA processing | | Descriptor: | 28-MER, MAGNESIUM ION, Ribonuclease III | | Authors: | Gan, J, Tropea, J.E, Austin, B.P, Court, D.L, Waugh, D.S, Ji, X. | | Deposit date: | 2005-11-10 | | Release date: | 2006-02-07 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural Insight into the Mechanism of Double-Stranded RNA Processing by Ribonuclease III.
Cell(Cambridge,Mass.), 124, 2006
|
|
2G1K
 
 | | Crystal structure of Mycobacterium tuberculosis shikimate kinase in complex with shikimate at 1.75 angstrom resolution | | Descriptor: | (3R,4S,5R)-3,4,5-TRIHYDROXYCYCLOHEX-1-ENE-1-CARBOXYLIC ACID, CHLORIDE ION, SULFATE ION, ... | | Authors: | Gan, J, Gu, Y, Li, Y, Yan, H, Ji, X. | | Deposit date: | 2006-02-14 | | Release date: | 2006-07-18 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal Structure of Mycobacterium tuberculosis Shikimate Kinase in Complex with Shikimic Acid and an ATP Analogue.
Biochemistry, 45, 2006
|
|
1YZ9
 
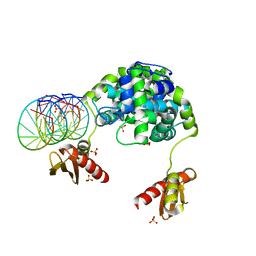 | | Crystal structure of RNase III mutant E110Q from Aquifex aeolicus complexed with double stranded RNA at 2.1-Angstrom Resolution | | Descriptor: | 5'-R(*CP*GP*AP*AP*CP*UP*UP*CP*GP*CP*G)-3', Ribonuclease III, SULFATE ION | | Authors: | Gan, J, Tropea, J.E, Austin, B.P, Court, D.L, Waugh, D.S, Ji, X. | | Deposit date: | 2005-02-28 | | Release date: | 2005-11-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Intermediate states of ribonuclease III in complex with double-stranded RNA
Structure, 13, 2005
|
|
1YYO
 
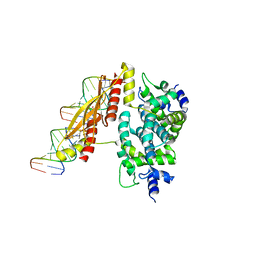 | | Crystal structure of RNase III mutant E110K from Aquifex aeolicus complexed with double-stranded RNA at 2.9-Angstrom Resolution | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 5'-R(*CP*GP*CP*GP*AP*AP*UP*UP*CP*GP*CP*G)-3', Ribonuclease III | | Authors: | Gan, J, Tropea, J.E, Austin, B.P, Court, D.L, Waugh, D.S, Ji, X. | | Deposit date: | 2005-02-25 | | Release date: | 2005-11-22 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Intermediate states of ribonuclease III in complex with double-stranded RNA
Structure, 13, 2005
|
|
1YYW
 
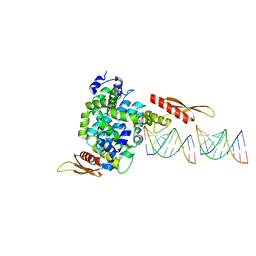 | | Crystal structure of RNase III from Aquifex aeolicus complexed with double stranded RNA at 2.8-Angstrom Resolution | | Descriptor: | 5'-R(*AP*AP*AP*UP*AP*UP*AP*UP*AP*UP*UP*U)-3', Ribonuclease III | | Authors: | Gan, J, Tropea, J.E, Austin, B.P, Court, D.L, Waugh, D.S, Ji, X. | | Deposit date: | 2005-02-25 | | Release date: | 2005-11-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Intermediate states of ribonuclease III in complex with double-stranded RNA
Structure, 13, 2005
|
|
1YYK
 
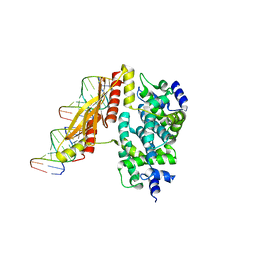 | | Crystal structure of RNase III from Aquifex Aeolicus complexed with double-stranded RNA at 2.5-angstrom resolution | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 5'-R(*CP*GP*CP*GP*AP*AP*UP*UP*CP*GP*CP*G)-3', Ribonuclease III | | Authors: | Gan, J, Tropea, J.E, Austin, B.P, Court, D.L, Waugh, D.S, Ji, X. | | Deposit date: | 2005-02-25 | | Release date: | 2005-11-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Intermediate states of ribonuclease III in complex with double-stranded RNA
Structure, 13, 2005
|
|
4M2Z
 
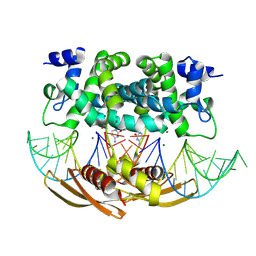 | | Crystal structure of RNASE III complexed with double-stranded RNA and CMP (TYPE II CLEAVAGE) | | Descriptor: | CYTIDINE-5'-MONOPHOSPHATE, MAGNESIUM ION, RNA10, ... | | Authors: | Gan, J, Liang, Y.-H, Shaw, G.X, Tropea, J.E, Waugh, D.S, Ji, X. | | Deposit date: | 2013-08-05 | | Release date: | 2013-12-11 | | Last modified: | 2024-07-03 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | RNase III: Genetics and Function; Structure and Mechanism.
Annu. Rev. Genet., 47, 2013
|
|
4M30
 
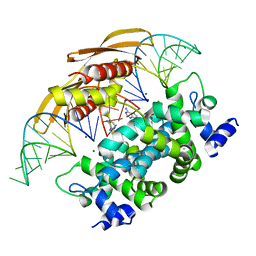 | | Crystal structure of RNASE III complexed with double-stranded RNA AND AMP (TYPE II CLEAVAGE) | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ADENOSINE MONOPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Gan, J, Liang, Y.-H, Shaw, G.X, Tropea, J.E, Waugh, D.S, Ji, X. | | Deposit date: | 2013-08-05 | | Release date: | 2013-12-11 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.501 Å) | | Cite: | RNase III: Genetics and Function; Structure and Mechanism.
Annu. Rev. Genet., 47, 2013
|
|
8CI0
 
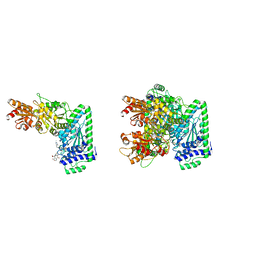 | | Maize Transketolase in complex with TPP and hydrolyzed (+)-Cornexistin | | Descriptor: | (1~{Z},3~{R},4~{S},7~{S},8~{Z})-8-ethylidene-4,7-bis(oxidanyl)-5-oxidanylidene-3-propyl-cyclononene-1,2-dicarboxylic acid, 2-[3-[(4-azanyl-2-methyl-pyrimidin-5-yl)methyl]-4-methyl-2H-1,3-thiazol-5-yl]ethyl phosphono hydrogen phosphate, MAGNESIUM ION, ... | | Authors: | Freigang, J. | | Deposit date: | 2023-02-08 | | Release date: | 2023-03-22 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.902 Å) | | Cite: | Investigations into Simplified Analogues of the Herbicidal Natural Product (+)-Cornexistin.
Chemistry, 29, 2023
|
|
6FBM
 
 | |
8P8K
 
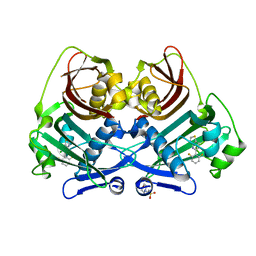 | | Acyl-ACP thioesterase from Lemna paucicostata in complex with a thiazolopyridine | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 5-[2,6-bis(fluoranyl)phenyl]-6-chloranyl-[1,3]thiazolo[4,5-b]pyridine, Acyl-ACP thioesterase | | Authors: | Freigang, J. | | Deposit date: | 2023-06-01 | | Release date: | 2023-09-20 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | A Study in Scaffold Hopping: Discovery and Optimization of Thiazolopyridines as Potent Herbicides That Inhibit Acyl-ACP Thioesterase.
J.Agric.Food Chem., 71, 2023
|
|
8QRT
 
 | |
8QS0
 
 | |
1FUE
 
 | | FLAVODOXIN FROM HELICOBACTER PYLORI | | Descriptor: | FLAVIN MONONUCLEOTIDE, FLAVODOXIN | | Authors: | Freigang, J, Diederichs, K, Schaefer, K.P, Welte, W, Paul, R. | | Deposit date: | 2000-09-15 | | Release date: | 2002-02-06 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of oxidized flavodoxin, an essential protein in Helicobacter pylori.
Protein Sci., 11, 2002
|
|
5OR2
 
 | | Crystal structures of PYR1/HAB1 in complex with synthetic analogues of Abscisic Acid | | Descriptor: | (2~{Z},4~{E})-3-cyclopropyl-5-[(1~{S})-2,6,6-trimethyl-1-oxidanyl-4-oxidanylidene-cyclohex-2-en-1-yl]penta-2,4-dienoic acid, Abscisic acid receptor PYR1, MANGANESE (II) ION, ... | | Authors: | Freigang, J. | | Deposit date: | 2017-08-15 | | Release date: | 2018-06-27 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Insights into the in Vitro and in Vivo SAR of Abscisic Acid - Exploring Unprecedented Variations of the Side Chain via Cross-Coupling-Mediated Syntheses
Eur.J.Org.Chem., 2018
|
|
5OR6
 
 | | Crystal structures of PYR1/HAB1 in complex with synthetic analogues of Abscisic Acid | | Descriptor: | (~{E})-3-(trifluoromethyl)-5-[(1~{S})-2,6,6-trimethyl-1-oxidanyl-4-oxidanylidene-cyclohex-2-en-1-yl]pent-2-en-4-ynoic acid, Abscisic acid receptor PYR1, MANGANESE (II) ION, ... | | Authors: | Freigang, J. | | Deposit date: | 2017-08-15 | | Release date: | 2018-06-27 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Insights into the in Vitro and in Vivo SAR of Abscisic Acid - Exploring Unprecedented Variations of the Side Chain via Cross-Coupling-Mediated Syntheses
Eur.J.Org.Chem., 2018
|
|
1CS6
 
 | | N-TERMINAL FRAGMENT OF AXONIN-1 FROM CHICKEN | | Descriptor: | AXONIN-1, GLYCEROL | | Authors: | Freigang, J, Proba, K, Diederichs, K, Sonderegger, P, Welte, W. | | Deposit date: | 1999-08-17 | | Release date: | 2000-05-19 | | Last modified: | 2017-10-04 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The crystal structure of the ligand binding module of axonin-1/TAG-1 suggests a zipper mechanism for neural cell adhesion.
Cell(Cambridge,Mass.), 101, 2000
|
|
3LTR
 
 | | 5-OMe-dU containing DNA 8mer | | Descriptor: | 5'-D(*GP*(UMS)P*GP*(T5O)P*AP*CP*AP*C)-3', MAGNESIUM ION | | Authors: | Sheng, J, Hassan, A.E.A, Zhang, W, Gan, J, Huang, Z. | | Deposit date: | 2010-02-16 | | Release date: | 2010-03-09 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Hydrogen bond formation between the naturally modified nucleobase and phosphate backbone.
Nucleic Acids Res., 40, 2012
|
|
6LRB
 
 | |
