7A98
 
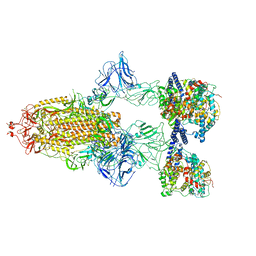 | | SARS-CoV-2 Spike Glycoprotein with 3 ACE2 Bound | | Descriptor: | Angiotensin-converting enzyme 2, Spike glycoprotein | | Authors: | Benton, D.J, Wrobel, A.G, Rosenthal, P.B, Gamblin, S.J. | | Deposit date: | 2020-09-01 | | Release date: | 2020-09-23 | | Last modified: | 2020-12-16 | | Method: | ELECTRON MICROSCOPY (5.4 Å) | | Cite: | Receptor binding and priming of the spike protein of SARS-CoV-2 for membrane fusion.
Nature, 588, 2020
|
|
7BNO
 
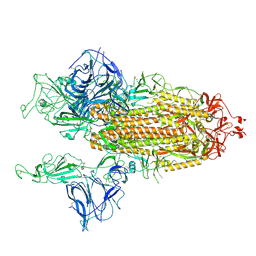 | | Open conformation of D614G SARS-CoV-2 spike with 2 Erect RBDs | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Benton, D.J, Wrobel, A.G, Rosenthal, P.B, Gamblin, S.J. | | Deposit date: | 2021-01-22 | | Release date: | 2021-02-03 | | Last modified: | 2021-03-10 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | The effect of the D614G substitution on the structure of the spike glycoprotein of SARS-CoV-2.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7BNM
 
 | | Closed conformation of D614G SARS-CoV-2 spike protein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Benton, D.J, Wrobel, A.G, Rosenthal, P.B, Gamblin, S.J. | | Deposit date: | 2021-01-22 | | Release date: | 2021-02-03 | | Last modified: | 2021-03-10 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | The effect of the D614G substitution on the structure of the spike glycoprotein of SARS-CoV-2.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7A5S
 
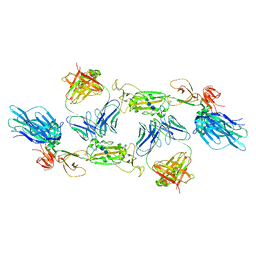 | | Complex of SARS-CoV-2 spike and CR3022 Fab (Homogeneous Refinement) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CR3022 Fab Heavy Chain, CR3022 Fab Light Chain, ... | | Authors: | Wrobel, A.G, Benton, D.J, Rosenthal, P.B, Gamblin, S.J. | | Deposit date: | 2020-08-21 | | Release date: | 2020-09-16 | | Last modified: | 2020-11-04 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Antibody-mediated disruption of the SARS-CoV-2 spike glycoprotein.
Nat Commun, 11, 2020
|
|
7A5R
 
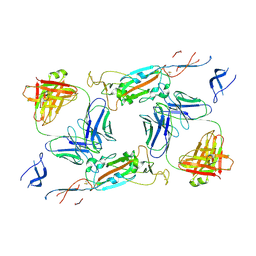 | | Complex of SARS-CoV-2 spike and CR3022 Fab (Non-Uniform Refinement) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CR3022 Fab Heavy Chain, CR3022 Fab Light Chain, ... | | Authors: | Wrobel, A.G, Benton, D.J, Rosenthal, P.B, Gamblin, S.J. | | Deposit date: | 2020-08-21 | | Release date: | 2020-09-16 | | Last modified: | 2020-11-04 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Antibody-mediated disruption of the SARS-CoV-2 spike glycoprotein.
Nat Commun, 11, 2020
|
|
7A95
 
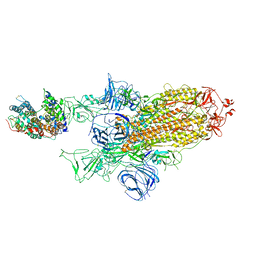 | | SARS-CoV-2 Spike Glycoprotein with 1 ACE2 Bound and 1 RBD Erect in Clockwise Direction | | Descriptor: | Angiotensin-converting enzyme 2, Spike glycoprotein | | Authors: | Benton, D.J, Wrobel, A.G, Rosenthal, P.B, Gamblin, S.J. | | Deposit date: | 2020-09-01 | | Release date: | 2020-09-16 | | Last modified: | 2020-12-16 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Receptor binding and priming of the spike protein of SARS-CoV-2 for membrane fusion.
Nature, 588, 2020
|
|
7A92
 
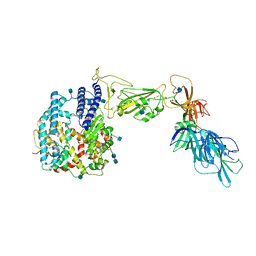 | | Dissociated S1 domain of SARS-CoV-2 Spike bound to ACE2 (Unmasked Refinement) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | Authors: | Benton, D.J, Wrobel, A.G, Rosenthal, P.B, Gamblin, S.J. | | Deposit date: | 2020-09-01 | | Release date: | 2020-09-30 | | Last modified: | 2020-12-16 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Receptor binding and priming of the spike protein of SARS-CoV-2 for membrane fusion.
Nature, 588, 2020
|
|
7A91
 
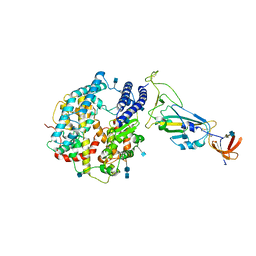 | | Dissociated S1 domain of SARS-CoV-2 Spike bound to ACE2 (Non-Uniform Refinement) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | Authors: | Benton, D.J, Wrobel, A.G, Rosenthal, P.B, Gamblin, S.J. | | Deposit date: | 2020-09-01 | | Release date: | 2020-09-16 | | Last modified: | 2020-12-16 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Receptor binding and priming of the spike protein of SARS-CoV-2 for membrane fusion.
Nature, 588, 2020
|
|
7A94
 
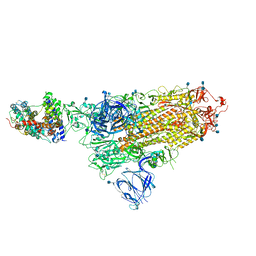 | | SARS-CoV-2 Spike Glycoprotein with 1 ACE2 Bound | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, Spike glycoprotein, ... | | Authors: | Benton, D.J, Wrobel, A.G, Rosenthal, P.B, Gamblin, S.J. | | Deposit date: | 2020-09-01 | | Release date: | 2020-09-16 | | Last modified: | 2020-12-16 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Receptor binding and priming of the spike protein of SARS-CoV-2 for membrane fusion.
Nature, 588, 2020
|
|
7A96
 
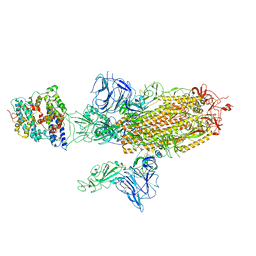 | | SARS-CoV-2 Spike Glycoprotein with 1 ACE2 Bound and 1 RBD Erect in Anticlockwise Direction | | Descriptor: | Angiotensin-converting enzyme 2, Spike glycoprotein | | Authors: | Benton, D.J, Wrobel, A.G, Rosenthal, P.B, Gamblin, S.J. | | Deposit date: | 2020-09-01 | | Release date: | 2020-09-16 | | Last modified: | 2020-12-16 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | Receptor binding and priming of the spike protein of SARS-CoV-2 for membrane fusion.
Nature, 588, 2020
|
|
7A97
 
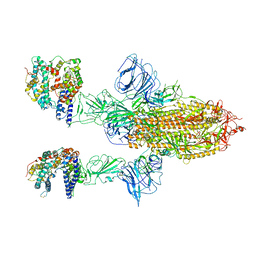 | | SARS-CoV-2 Spike Glycoprotein with 2 ACE2 Bound | | Descriptor: | Angiotensin-converting enzyme 2, Spike glycoprotein | | Authors: | Benton, D.J, Wrobel, A.G, Rosenthal, P.B, Gamblin, S.J. | | Deposit date: | 2020-09-01 | | Release date: | 2020-09-16 | | Last modified: | 2020-12-16 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Receptor binding and priming of the spike protein of SARS-CoV-2 for membrane fusion.
Nature, 588, 2020
|
|
7A93
 
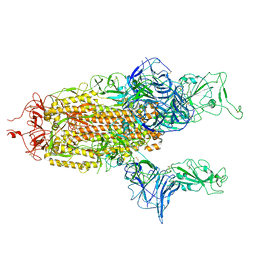 | |
1AM4
 
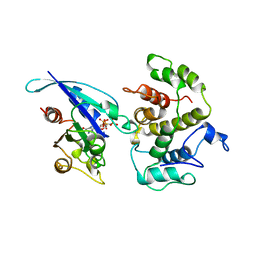 | | COMPLEX BETWEEN CDC42HS.GMPPNP AND P50 RHOGAP (H. SAPIENS) | | Descriptor: | CDC42HS, MAGNESIUM ION, P50-RHOGAP, ... | | Authors: | Rittinger, K, Walker, P, Gamblin, S.J, Smerdon, S.J. | | Deposit date: | 1997-06-22 | | Release date: | 1998-07-15 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of a small G protein in complex with the GTPase-activating protein rhoGAP.
Nature, 388, 1997
|
|
5IQP
 
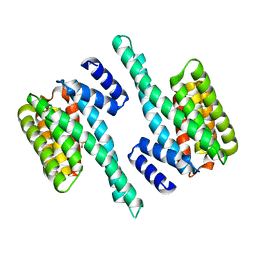 | | 14-3-3 PROTEIN TAU ISOFORM | | Descriptor: | 14-3-3 protein theta, SULFATE ION | | Authors: | Xiao, B, Smerdon, S.J, Gamblin, S.J. | | Deposit date: | 2016-03-11 | | Release date: | 2016-03-23 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.602 Å) | | Cite: | Structure of a 14-3-3 protein and implications for coordination of multiple signalling pathways
Nature, 376, 1995
|
|
5AJM
 
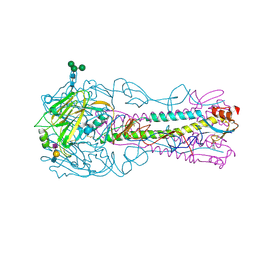 | | H5 (VN1194) Asn186Lys Mutant Haemagglutinin in Complex with Avian Receptor Analogue 3'SLN | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 3[N-MORPHOLINO]PROPANE SULFONIC ACID, ... | | Authors: | Xiong, X, Xiao, H, Martin, S.R, Coombs, P.J, Liu, J, Collins, P.J, Vachieri, S.G, Walker, P.A, Lin, Y.P, McCauley, J.W, Gamblin, S.J, Skehel, J.J. | | Deposit date: | 2015-02-25 | | Release date: | 2015-03-25 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Enhanced Human Receptor Binding by H5 Haemagglutinins.
Virology, 456, 2014
|
|
5A3I
 
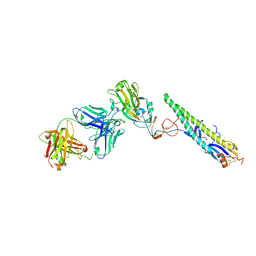 | | Crystal Structure of a Complex formed between FLD194 Fab and Transmissible Mutant H5 Haemagglutinin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ANTI-HAEMAGGLUTININ HA1 FAB HEAVY CHAIN, ... | | Authors: | Xiong, X, Corti, D, Liu, J, Pinna, D, Foglierini, M, Calder, L.J, Martin, S.R, Lin, Y.P, Walker, P.A, Collins, P.J, Monne, I, Suguitan Jr, A.L, Santos, C, Temperton, N.J, Subbarao, K, Lanzavecchia, A, Gamblin, S.J, Skehel, J.J. | | Deposit date: | 2015-06-01 | | Release date: | 2015-07-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.89 Å) | | Cite: | Structures of Complexes Formed by H5 Influenza Hemagglutinin with a Potent Broadly Neutralizing Human Monoclonal Antibody.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
2BQZ
 
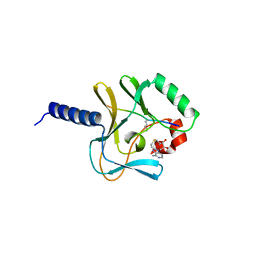 | | Crystal structure of a ternary complex of the human histone methyltransferase Pr-SET7 (also known as SET8) | | Descriptor: | HISTONE H4, S-ADENOSYL-L-HOMOCYSTEINE, SET8 PROTEIN | | Authors: | Xiao, B, Jing, C, Kelly, G, Walker, P.A, Muskett, F.W, Frenkiel, T.A, Martin, S.R, Sarma, K, Reinberg, D, Gamblin, S.J, Wilson, J.R. | | Deposit date: | 2005-04-28 | | Release date: | 2005-06-08 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Specificity and Mechanism of the Histone Methyltransferase Pr-Set7
Genes Dev., 19, 2005
|
|
2AIO
 
 | | Metallo beta lactamase L1 from Stenotrophomonas maltophilia complexed with hydrolyzed moxalactam | | Descriptor: | (2R)-2-((R)-CARBOXY{[CARBOXY(4-HYDROXYPHENYL)ACETYL]AMINO}METHOXYMETHYL)-5-METHYLENE-5,6-DIHYDRO-2H-1,3-OXAZINE-4-CARBO XYLIC ACID, Metallo-beta-lactamase L1, SULFATE ION, ... | | Authors: | Spencer, J, Read, J, Sessions, R.B, Howell, S, Blackburn, G.M, Gamblin, S.J. | | Deposit date: | 2005-07-30 | | Release date: | 2005-10-11 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Antibiotic Recognition by Binuclear Metallo-beta-Lactamases Revealed by X-ray Crystallography
J.Am.Chem.Soc., 127, 2005
|
|
3T4N
 
 | | Structure of the regulatory fragment of Saccharomyces cerevisiae AMPK in complex with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Carbon catabolite-derepressing protein kinase, Nuclear protein SNF4, ... | | Authors: | Mayer, F.V, Heath, R, Underwood, E, Sanders, M.J, Carmena, D, McCartney, R, Leiper, F.C, Xiao, B, Jing, C, Walker, P.A, Haire, L.F, Ogrodowicz, R, Martin, S.R, Schmdit, M.C, Gamblin, S.J, Carling, D. | | Deposit date: | 2011-07-26 | | Release date: | 2011-11-09 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | ADP Regulates SNF1, the Saccharomyces cerevisiae Homolog of AMP-Activated Protein Kinase.
Cell Metab, 14, 2011
|
|
1MXE
 
 | | Structure of the Complex of Calmodulin with the Target Sequence of CaMKI | | Descriptor: | CALCIUM ION, Calmodulin, Target Sequence of rat Calmodulin-Dependent Protein Kinase I | | Authors: | Clapperton, J.A, Martin, S.R, Smerdon, S.J, Gamblin, S.J, Bayley, P.M. | | Deposit date: | 2002-10-02 | | Release date: | 2002-12-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of the Complex
of Calmodulin with the Target
Sequence of Calmodulin-Dependent
Protein Kinase I: Studies of the
Kinase Activation Mechanism
Biochemistry, 41, 2002
|
|
1OW3
 
 | | Crystal Structure of RhoA.GDP.MgF3-in Complex with RhoGAP | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Rho-GTPase-activating protein 1, ... | | Authors: | Graham, D.L, Lowe, P.N, Grime, G.W, Marsh, M, Rittinger, K, Smerdon, S.J, Gamblin, S.J, Eccleston, J.F. | | Deposit date: | 2003-03-28 | | Release date: | 2003-05-06 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | MgF(3)(-) as a Transition State Analog of Phosphoryl Transfer
Chem.Biol., 9, 2002
|
|
3TDH
 
 | | Structure of the regulatory fragment of sccharomyces cerevisiae AMPK in complex with AMP | | Descriptor: | ADENOSINE MONOPHOSPHATE, Carbon catabolite-derepressing protein kinase, Nuclear protein SNF4, ... | | Authors: | Mayer, F.V, Heath, R, Underwood, E, Sanders, M.J, Carmena, D, McCartney, R, Leiper, F.C, Xiao, B, Jing, C, Walker, P.A, Haire, L.F, Ogrodowicz, R, Martin, S.R, Schmidt, M.C, Gamblin, S.J, Carling, D. | | Deposit date: | 2011-08-11 | | Release date: | 2011-11-09 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | ADP Regulates SNF1, the Saccharomyces cerevisiae Homolog of AMP-Activated Protein Kinase.
Cell Metab, 14, 2011
|
|
1QJB
 
 | | 14-3-3 ZETA/PHOSPHOPEPTIDE COMPLEX (MODE 1) | | Descriptor: | 14-3-3 PROTEIN ZETA/DELTA, PHOSPHOPEPTIDE | | Authors: | Rittinger, K, Budman, J, Xu, J, Volinia, S, Cantley, L.C, Smerdon, S.J, Gamblin, S.J, Yaffe, M.B. | | Deposit date: | 1999-06-23 | | Release date: | 1999-09-15 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Analysis of 14-3-3 Phosphopeptide Complexes Identifies a Dual Role for the Nuclear Export Signal of 14-3-3 in Ligand Binding
Mol.Cell, 4, 1999
|
|
3TE5
 
 | | structure of the regulatory fragment of sacchromyces cerevisiae ampk in complex with NADH | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, Carbon catabolite-derepressing protein kinase, Nuclear protein SNF4, ... | | Authors: | Mayer, F.V, Heath, R, Underwood, E, Sanders, M.J, Carmena, D, McCartney, R, Leiper, F.C, Xiao, B, Jing, C, Walker, P.A, Haire, L.F, Ogrodowicz, R, Martin, S.R, Schmidt, M.C, Gamblin, S.J, Carling, D. | | Deposit date: | 2011-08-12 | | Release date: | 2011-11-09 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | ADP Regulates SNF1, the Saccharomyces cerevisiae Homolog of AMP-Activated Protein Kinase.
Cell Metab, 14, 2011
|
|
1QJA
 
 | | 14-3-3 ZETA/PHOSPHOPEPTIDE COMPLEX (MODE 2) | | Descriptor: | 14-3-3 PROTEIN ZETA, PHOSPHOPEPTIDE | | Authors: | Rittinger, K, Budman, J, Xu, J, Volinia, S, Cantley, L.C, Smerdon, S.J, Gamblin, S.J, Yaffe, M.B. | | Deposit date: | 1999-06-23 | | Release date: | 1999-09-15 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Analysis of 14-3-3 Phosphopeptide Complexes Identifies a Dual Role for the Nuclear Export Signal of 14-3-3 in Ligand Binding
Mol.Cell, 4, 1999
|
|
