5U9I
 
 | | Crystal structure of the FKBP domain of human aryl hydrocarbon receptor-interacting protein-like 1 (AIPL1) complexed with S-farnesyl-L-cysteine methyl ester | | Descriptor: | Aryl hydrocarbon receptor-interacting protein-like 1 (AIPL1), FARNESYL | | Authors: | Yadav, R.P, Gakhar, L, Liping, Y, Artemyev, N.O. | | Deposit date: | 2016-12-16 | | Release date: | 2017-07-26 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Unique structural features of the AIPL1-FKBP domain that support prenyl lipid binding and underlie protein malfunction in blindness.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5V35
 
 | | Crystal structure of V71F mutant of the FKBP domain of human aryl hydrocarbon receptor-interacting protein-like 1 (AIPL1) complexed with S-farnesyl-L-cysteine methyl ester | | Descriptor: | Aryl hydrocarbon receptor-interacting protein-like 1 (AIPL1), FARNESYL | | Authors: | Yadav, R.P, Gakhar, L, Liping, Y, Artemyev, N.O. | | Deposit date: | 2017-03-06 | | Release date: | 2017-07-26 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Unique structural features of the AIPL1-FKBP domain that support prenyl lipid binding and underlie protein malfunction in blindness.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5U9K
 
 | |
5U9A
 
 | |
5U9J
 
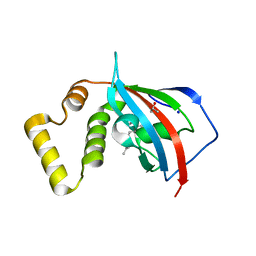 | | Crystal structure of the FKBP domain of human aryl hydrocarbon receptor-interacting protein-like 1 (AIPL1) complexed with geranyl geranyl pyrophoshate | | Descriptor: | Aryl hydrocarbon receptor-interacting protein-like 1 (AIPL1), GERAN-8-YL GERAN, ISOPROPYL ALCOHOL, ... | | Authors: | Yadav, R.P, Gakhar, L, Liping, Y, Artemyev, N.O. | | Deposit date: | 2016-12-16 | | Release date: | 2017-07-26 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Unique structural features of the AIPL1-FKBP domain that support prenyl lipid binding and underlie protein malfunction in blindness.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
7UHW
 
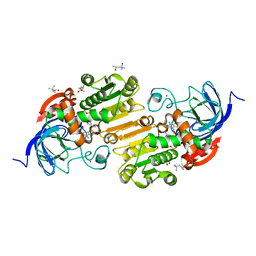 | | Horse liver alcohol dehydrogenase G173A, complexed with NAD+ and 2,3,4,5,6-pentafluorobenzyl alcohol at 120 K | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, 2,3,4,5,6-PENTAFLUOROBENZYL ALCOHOL, Alcohol dehydrogenase E chain, ... | | Authors: | Plapp, B.V, Gakhar, L. | | Deposit date: | 2022-03-27 | | Release date: | 2022-04-06 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Dependence of crystallographic atomic displacement factors on temperature (25-150 K) for complexes of horse liver alcohol dehydrogenases
Acta Crystallogr.,Sect.D, D78, 2022
|
|
7UHX
 
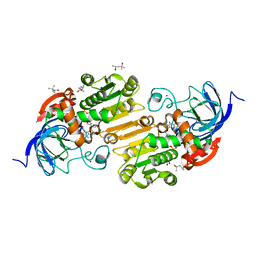 | | Horse liver alcohol dehydrogenase G173A, complexed with NAD+ and 2,3,4,5,6-pentafluorobenzyl alcohol at 150 K | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, 2,3,4,5,6-PENTAFLUOROBENZYL ALCOHOL, Alcohol dehydrogenase E chain, ... | | Authors: | Plapp, B.V, Gakhar, L. | | Deposit date: | 2022-03-27 | | Release date: | 2022-04-06 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Dependence of crystallographic atomic displacement factors on temperature (25-150 K) for complexes of horse liver alcohol dehydrogenases
Acta Crystallogr.,Sect.D, D78, 2022
|
|
7UHV
 
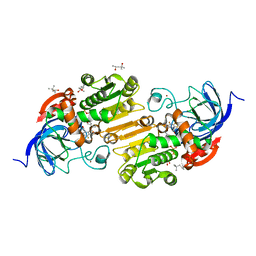 | | Horse liver alcohol dehydrogenase G173A, complexed with NAD+ and 2,3,4,5,6-pentafluorobenzyl alcohol at 50 K | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, 2,3,4,5,6-PENTAFLUOROBENZYL ALCOHOL, Alcohol dehydrogenase E chain, ... | | Authors: | Plapp, B.V, Gakhar, L. | | Deposit date: | 2022-03-27 | | Release date: | 2022-04-06 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Dependence of crystallographic atomic displacement factors on temperature (25-150 K) for complexes of horse liver alcohol dehydrogenases
Acta Crystallogr.,Sect.D, D78, 2022
|
|
4JIO
 
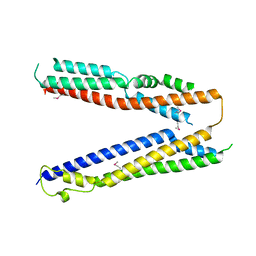 | | Bro1 V domain and ubiquitin | | Descriptor: | BRO1, Ubiquitin | | Authors: | Pashkova, N, Gakhar, L, Piper, R.C. | | Deposit date: | 2013-03-06 | | Release date: | 2013-06-19 | | Last modified: | 2013-07-10 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | The yeast alix homolog bro1 functions as a ubiquitin receptor for protein sorting into multivesicular endosomes.
Dev.Cell, 25, 2013
|
|
4JJY
 
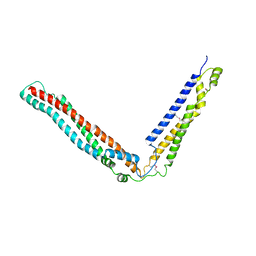 | | Alix V domain | | Descriptor: | Programmed cell death 6-interacting protein | | Authors: | Pashkova, N, Gakhar, L, Yu, L, Piper, R.C. | | Deposit date: | 2013-03-08 | | Release date: | 2013-06-19 | | Last modified: | 2013-07-10 | | Method: | X-RAY DIFFRACTION (6.503 Å) | | Cite: | The yeast alix homolog bro1 functions as a ubiquitin receptor for protein sorting into multivesicular endosomes.
Dev.Cell, 25, 2013
|
|
4QLF
 
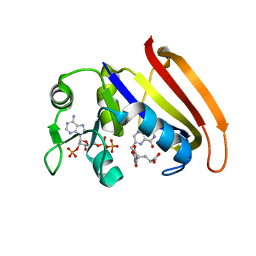 | |
4QLE
 
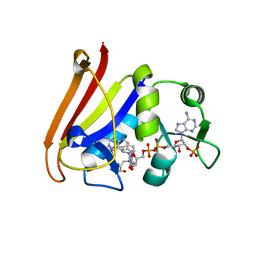 | |
4QLG
 
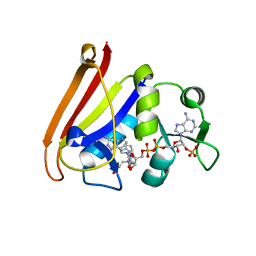 | |
4K2P
 
 | |
4ID3
 
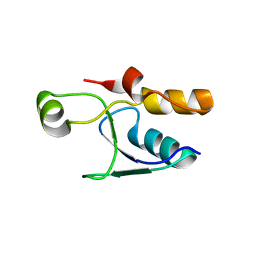 | |
6N85
 
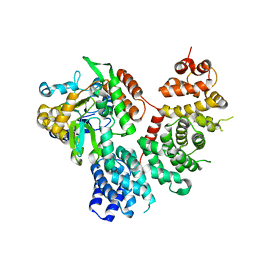 | | Resistance to inhibitors of cholinesterase 8A (Ric8A) protein in complex with MBP-tagged transducin-alpha residues 327-350 | | Descriptor: | Maltose/maltodextrin-binding periplasmic protein,Guanine nucleotide-binding protein G(t) subunit alpha-2, Synembryn-A, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Srivastava, D, Gakhar, L, Artemyev, N.O. | | Deposit date: | 2018-11-28 | | Release date: | 2019-07-10 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural underpinnings of Ric8A function as a G-protein alpha-subunit chaperone and guanine-nucleotide exchange factor.
Nat Commun, 10, 2019
|
|
4K2O
 
 | |
6N84
 
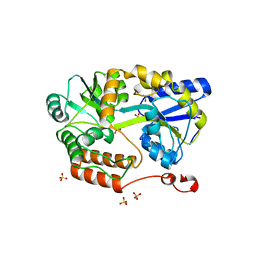 | | MBP-fusion protein of transducin-alpha residues 327-350 | | Descriptor: | Maltose/maltodextrin-binding periplasmic protein,Guanine nucleotide-binding protein G(t) subunit alpha-2, SULFATE ION, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Srivastava, D, Gakhar, L, Artemyev, N.O. | | Deposit date: | 2018-11-28 | | Release date: | 2019-07-10 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural underpinnings of Ric8A function as a G-protein alpha-subunit chaperone and guanine-nucleotide exchange factor.
Nat Commun, 10, 2019
|
|
6N86
 
 | |
4JQW
 
 | | Crystal Structure of a Complex of NOD1 CARD and Ubiquitin | | Descriptor: | Nucleotide-binding oligomerization domain-containing protein 1, PHOSPHATE ION, Polyubiquitin-C | | Authors: | Ver Heul, A.M, Gakhar, L, Piper, R.C, Ramaswamy, S. | | Deposit date: | 2013-03-20 | | Release date: | 2014-03-26 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of a complex of NOD1 CARD and ubiquitin
Plos One, 9, 2014
|
|
5V6Z
 
 | |
5V6U
 
 | |
7UA6
 
 | | Horse liver alcohol dehydrogenase with NAD and trifluoroethanol at 25K | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, Alcohol dehydrogenase E chain, NICOTINAMIDE-ADENINE-DINUCLEOTIDE (ACIDIC FORM), ... | | Authors: | Plapp, B.V, Gakhar, L. | | Deposit date: | 2022-03-11 | | Release date: | 2022-03-23 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Dependence of crystallographic atomic displacement factors on temperature (25-150 K) for complexes of horse liver alcohol dehydrogenases
Acta Crystallogr.,Sect.D, D78, 2022
|
|
7UEI
 
 | | Horse liver alcohol dehydrogenase with NAD and pentafluorobenzyl alcohol at 100 K, 1.2 A, crystal 16 | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, 2,3,4,5,6-PENTAFLUOROBENZYL ALCOHOL, Alcohol dehydrogenase E chain, ... | | Authors: | Plapp, B.V, Gakhar, L. | | Deposit date: | 2022-03-21 | | Release date: | 2022-03-30 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Dependence of crystallographic atomic displacement factors on temperature (25-150 K) for complexes of horse liver alcohol dehydrogenases
Acta Crystallogr.,Sect.D, D78, 2022
|
|
7UC9
 
 | | Horse liver alcohol dehydrogenase with NAD and trifluoroethanol at 45 K | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, Alcohol dehydrogenase E chain, NICOTINAMIDE-ADENINE-DINUCLEOTIDE (ACIDIC FORM), ... | | Authors: | Plapp, B.V, Gakhar, L. | | Deposit date: | 2022-03-16 | | Release date: | 2022-03-30 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Dependence of crystallographic atomic displacement factors on temperature (25-150 K) for complexes of horse liver alcohol dehydrogenases
Acta Crystallogr.,Sect.D, D78, 2022
|
|
