7MMV
 
 | |
7MMP
 
 | |
7MMS
 
 | |
7MMR
 
 | |
7MMU
 
 | |
7MMQ
 
 | |
7MMW
 
 | |
2LV6
 
 | | The complex between Ca-Calmodulin and skeletal muscle myosin light chain kinase from combination of NMR and aqueous and contrast-matched SAXS data | | Descriptor: | CALCIUM ION, Calmodulin, Myosin light chain kinase 2, ... | | Authors: | Grishaev, A.V, Anthis, N.J, Clore, G.M. | | Deposit date: | 2012-06-29 | | Release date: | 2013-02-20 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR, SOLUTION SCATTERING | | Cite: | Contrast-matched small-angle X-ray scattering from a heavy-atom-labeled protein in structure determination: application to a lead-substituted calmodulin-peptide complex.
J.Am.Chem.Soc., 134, 2012
|
|
2MJH
 
 | |
5ZHQ
 
 | |
8SSH
 
 | | MtrR from Neisseria gonorrhoeae bound to Ethinyl Estradiol | | Descriptor: | Ethinyl estradiol, HTH-type transcriptional regulator MtrR, PHOSPHATE ION | | Authors: | Hooks, G.M, Brennan, R.G. | | Deposit date: | 2023-05-08 | | Release date: | 2024-02-14 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Hormonal steroids induce multidrug resistance and stress response genes in Neisseria gonorrhoeae by binding to MtrR.
Nat Commun, 15, 2024
|
|
2AXV
 
 | | Structure of PrgX Y153C mutant | | Descriptor: | PrgX | | Authors: | Shi, K, Brown, C.K, Gu, Z.Y, Kozlowicz, B.K, Dunny, G.M, Ohlendorf, D.H, Earhart, C.A. | | Deposit date: | 2005-09-06 | | Release date: | 2005-12-06 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure of peptide sex pheromone receptor PrgX and PrgX/pheromone complexes and regulation of conjugation in Enterococcus faecalis.
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
2AZB
 
 | | HIV-1 Protease NL4-3 3X mutant in complex with inhibitor, TL-3 | | Descriptor: | PROTEASE RETROPEPSIN, benzyl [(1S,4S,7S,8R,9R,10S,13S,16S)-7,10-dibenzyl-8,9-dihydroxy-1,16-dimethyl-4,13-bis(1-methylethyl)-2,5,12,15,18-pentaoxo-20-phenyl-19-oxa-3,6,11,14,17-pentaazaicos-1-yl]carbamate | | Authors: | Heaslet, H, Kutilek, V, Morris, G.M, Lin, Y.-C, Elder, J.H, Torbett, B.E, Stout, C.D. | | Deposit date: | 2005-09-09 | | Release date: | 2006-02-28 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Structural Insights into the Mechanisms of Drug Resistance in HIV-1 Protease NL4-3
J.Mol.Biol., 356, 2006
|
|
2AS9
 
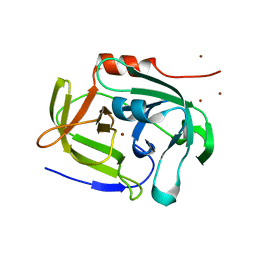 | | Functional and structural characterization of Spl proteases from staphylococcus aureus | | Descriptor: | ZINC ION, serine protease | | Authors: | Popowicz, G.M, Dubin, G, Stec-Niemczyk, J, Czarny, A, Dubin, A, Potempa, J, Holak, T.A. | | Deposit date: | 2005-08-23 | | Release date: | 2005-09-06 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Functional and Structural Characterization of Spl Proteases from Staphylococcus aureus
J.Mol.Biol., 358, 2006
|
|
2AZ9
 
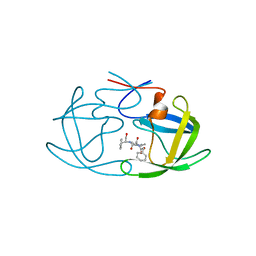 | | HIV-1 Protease NL4-3 1X mutant | | Descriptor: | PROTEASE RETROPEPSIN, benzyl [(1S,4S,7S,8R,9R,10S,13S,16S)-7,10-dibenzyl-8,9-dihydroxy-1,16-dimethyl-4,13-bis(1-methylethyl)-2,5,12,15,18-pentaoxo-20-phenyl-19-oxa-3,6,11,14,17-pentaazaicos-1-yl]carbamate | | Authors: | Heaslet, H, Kutilek, V, Morris, G.M, Lin, Y.-C, Elder, J.H, Torbett, B.E, Stout, C.D. | | Deposit date: | 2005-09-09 | | Release date: | 2006-02-28 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Insights into the Mechanisms of Drug Resistance in HIV-1 Protease NL4-3
J.Mol.Biol., 356, 2006
|
|
2AIO
 
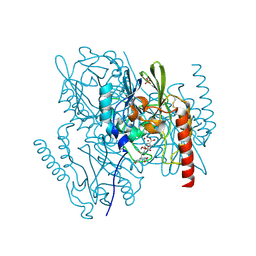 | | Metallo beta lactamase L1 from Stenotrophomonas maltophilia complexed with hydrolyzed moxalactam | | Descriptor: | (2R)-2-((R)-CARBOXY{[CARBOXY(4-HYDROXYPHENYL)ACETYL]AMINO}METHOXYMETHYL)-5-METHYLENE-5,6-DIHYDRO-2H-1,3-OXAZINE-4-CARBO XYLIC ACID, Metallo-beta-lactamase L1, SULFATE ION, ... | | Authors: | Spencer, J, Read, J, Sessions, R.B, Howell, S, Blackburn, G.M, Gamblin, S.J. | | Deposit date: | 2005-07-30 | | Release date: | 2005-10-11 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Antibiotic Recognition by Binuclear Metallo-beta-Lactamases Revealed by X-ray Crystallography
J.Am.Chem.Soc., 127, 2005
|
|
2AWI
 
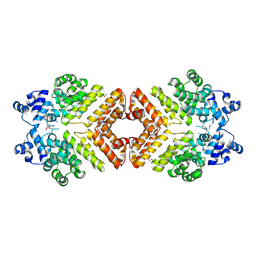 | | Structure of PrgX Y153C mutant | | Descriptor: | PrgX | | Authors: | Shi, K, Brown, C.K, Gu, Z.Y, Kozlowicz, B.k, Dunny, G.M, Ohlendorf, D.H, Earhart, C.A. | | Deposit date: | 2005-09-01 | | Release date: | 2005-12-06 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structure of peptide sex pheromone receptor PrgX and PrgX/pheromone complexes and regulation of conjugation in Enterococcus faecalis.
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
2AXU
 
 | | Structure of PrgX | | Descriptor: | PrgX | | Authors: | Shi, K, Brown, C.K, Gu, Z.Y, Kozlowicz, B.K, Dunny, G.M, Ohlendorf, D.H, Earhart, C.A. | | Deposit date: | 2005-09-06 | | Release date: | 2005-12-06 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure of peptide sex pheromone receptor PrgX and PrgX/pheromone complexes and regulation of conjugation in Enterococcus faecalis.
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
2AW6
 
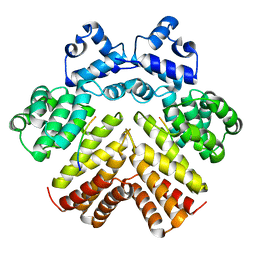 | | Structure of a bacterial peptide pheromone/receptor complex and its mechanism of gene regulation | | Descriptor: | PrgX, peptide | | Authors: | Shi, K, Brown, C.K, Gu, Z.Y, Kozlowicz, B.K, Dunny, G.M, Ohlendorf, D.H, Earhart, C.A. | | Deposit date: | 2005-08-31 | | Release date: | 2005-12-06 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure of peptide sex pheromone receptor PrgX and PrgX/pheromone complexes and regulation of conjugation in Enterococcus faecalis.
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
2AZ8
 
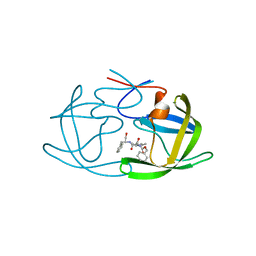 | | HIV-1 Protease NL4-3 in complex with inhibitor, TL-3 | | Descriptor: | PROTEASE RETROPEPSIN, benzyl [(1S,4S,7S,8R,9R,10S,13S,16S)-7,10-dibenzyl-8,9-dihydroxy-1,16-dimethyl-4,13-bis(1-methylethyl)-2,5,12,15,18-pentaoxo-20-phenyl-19-oxa-3,6,11,14,17-pentaazaicos-1-yl]carbamate | | Authors: | Heaslet, H, Kutilek, V, Morris, G.M, Lin, Y.-C, Elder, J.H, Torbett, B.E, Stout, C.D. | | Deposit date: | 2005-09-09 | | Release date: | 2006-02-28 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Insights into the Mechanisms of Drug Resistance in HIV-1 Protease NL4-3
J.Mol.Biol., 356, 2006
|
|
8QAO
 
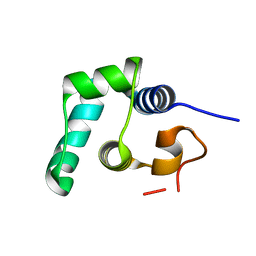 | |
1QLQ
 
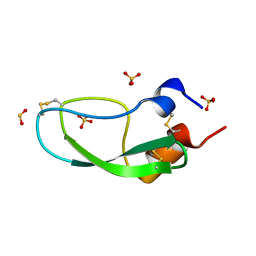 | | Bovine Pancreatic Trypsin Inhibitor (BPTI) Mutant with Altered Binding Loop Sequence | | Descriptor: | PANCREATIC TRYPSIN INHIBITOR, SULFATE ION | | Authors: | Czapinska, H, Krzywda, S, Sheldrick, G.M, Otlewski, J, Jaskolski, M. | | Deposit date: | 1999-09-10 | | Release date: | 1999-10-05 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | High Resolution Structure of Bovine Pancreatic Trypsin Inhibitor with Altered Binding Loop Sequence
J.Mol.Biol., 295, 1999
|
|
1R4T
 
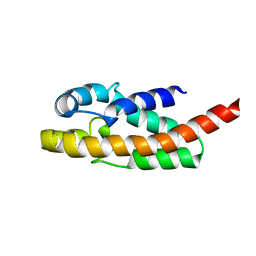 | | Solution structure of exoenzyme S | | Descriptor: | exoenzyme S | | Authors: | Langdon, G.M, Leitner, D, Labudde, D, Kuhne, R, Schmieder, P, Aktories, K, Oschkinat, H.O, Schmidt, G. | | Deposit date: | 2003-10-08 | | Release date: | 2005-04-12 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the N-terminal GTPase activating domain of Pseudomonas aeruginosa exoenzyme S
To be Published
|
|
1R36
 
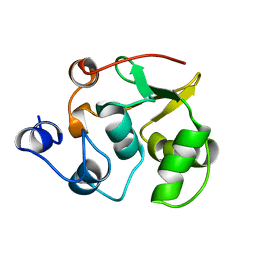 | | NMR-based structure of autoinhibited murine Ets-1 deltaN301 | | Descriptor: | C-ets-1 protein | | Authors: | Lee, G.M, Donaldson, L.W, Pufall, M.A, Kang, H.-S, Pot, I, Graves, B.J, McIntosh, L.P. | | Deposit date: | 2003-09-30 | | Release date: | 2004-11-09 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | The Structural and Dynamic Basis of Ets-1 DNA Binding Autoinhibition
J.Biol.Chem., 280, 2005
|
|
1RQW
 
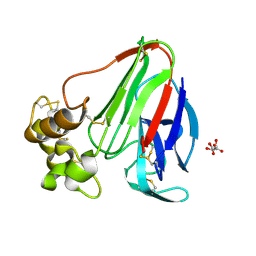 | |
