1KOA
 
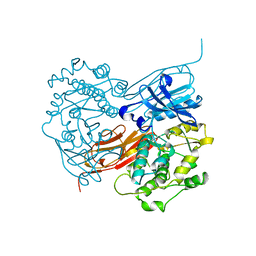 | | TWITCHIN KINASE FRAGMENT (C.ELEGANS), AUTOREGULATED PROTEIN KINASE AND IMMUNOGLOBULIN DOMAINS | | Descriptor: | TWITCHIN | | Authors: | Kobe, B, Heierhorst, J, Feil, S.C, Parker, M.W, Benian, G.M, Weiss, K.R, Kemp, B.E. | | Deposit date: | 1996-06-28 | | Release date: | 1997-03-12 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Giant protein kinases: domain interactions and structural basis of autoregulation.
EMBO J., 15, 1996
|
|
1KOB
 
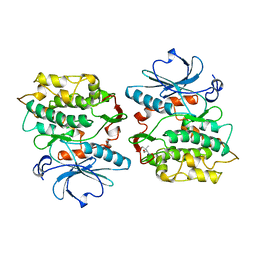 | | TWITCHIN KINASE FRAGMENT (APLYSIA), AUTOREGULATED PROTEIN KINASE DOMAIN | | Descriptor: | TWITCHIN, VALINE | | Authors: | Kobe, B, Heierhorst, J, Feil, S.C, Parker, M.W, Benian, G.M, Weiss, K.R, Kemp, B.E. | | Deposit date: | 1996-06-28 | | Release date: | 1997-03-12 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Giant protein kinases: domain interactions and structural basis of autoregulation.
EMBO J., 15, 1996
|
|
1N9K
 
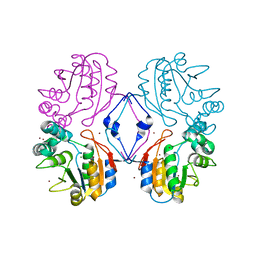 | | Crystal structure of the bromide adduct of AphA class B acid phosphatase/phosphotransferase from E. coli at 2.2 A resolution | | Descriptor: | BROMIDE ION, Class B acid phosphatase, MAGNESIUM ION | | Authors: | Calderone, V, Forleo, C, Benvenuti, M, Rossolini, G.M, Thaller, M.C, Mangani, S. | | Deposit date: | 2002-11-25 | | Release date: | 2004-02-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The first structure of a bacterial class B Acid phosphatase reveals further structural heterogeneity among phosphatases of the haloacid dehalogenase fold.
J.Mol.Biol., 335, 2004
|
|
1NER
 
 | |
1O15
 
 | |
1INY
 
 | | A SIALIC ACID DERIVED PHOSPHONATE ANALOG INHIBITS DIFFERENT STRAINS OF INFLUENZA VIRUS NEURAMINIDASE WITH DIFFERENT EFFICIENCIES | | Descriptor: | (1R)-4-acetamido-1,5-anhydro-2,4-dideoxy-1-phosphono-D-glycero-D-galacto-octitol, 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | White, C.L, Janakiraman, M.N, Laver, W.G, Philippon, C, Vasella, A, Air, G.M, Luo, M. | | Deposit date: | 1994-09-26 | | Release date: | 1995-02-07 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | A sialic acid-derived phosphonate analog inhibits different strains of influenza virus neuraminidase with different efficiencies.
J.Mol.Biol., 245, 1995
|
|
1QCK
 
 | |
1J47
 
 | |
1C1Z
 
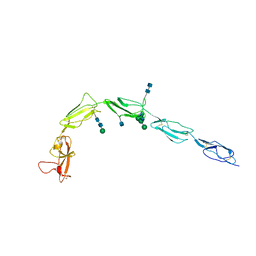 | | CRYSTAL STRUCTURE OF HUMAN BETA-2-GLYCOPROTEIN-I (APOLIPOPROTEIN-H) | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, BETA2-GLYCOPROTEIN-I, ... | | Authors: | Schwarzenbacher, R, Zeth, K, Diederichs, K, Gries, A, Kostner, G.M, Laggner, P, Prassl, R. | | Deposit date: | 1999-07-22 | | Release date: | 1999-11-19 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.87 Å) | | Cite: | Crystal structure of human beta2-glycoprotein I: implications for phospholipid binding and the antiphospholipid syndrome.
EMBO J., 18, 1999
|
|
2CM5
 
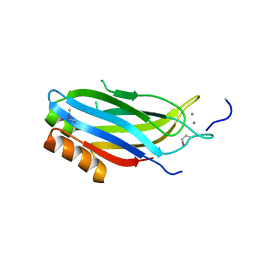 | | crystal structure of the C2B domain of rabphilin | | Descriptor: | CALCIUM ION, RABPHILIN-3A | | Authors: | Schlicker, C, Montaville, P, Sheldrick, G.M, Becker, S. | | Deposit date: | 2006-05-04 | | Release date: | 2006-12-04 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | The C2A-C2B Linker Defines the High Affinity Ca2+ Binding Mode of Rabphilin-3A.
J.Biol.Chem., 282, 2007
|
|
2DSQ
 
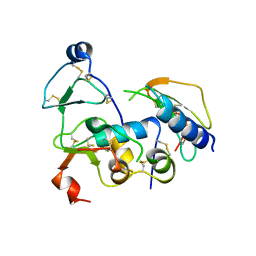 | | Structural Basis for the Inhibition of Insulin-like Growth Factors by IGF Binding Proteins | | Descriptor: | Insulin-like growth factor IB, Insulin-like growth factor-binding protein 1, Insulin-like growth factor-binding protein 4 | | Authors: | Sitar, T, Popowicz, G.M, Siwanowicz, I, Huber, R, Holak, T.A. | | Deposit date: | 2006-07-05 | | Release date: | 2006-08-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis for the inhibition of insulin-like growth factors by insulin-like growth factor-binding proteins.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
2DSP
 
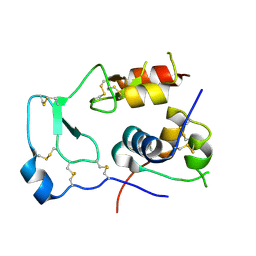 | | Structural Basis for the Inhibition of Insulin-like Growth Factors by IGF Binding Proteins | | Descriptor: | Insulin-like growth factor IB, Insulin-like growth factor-binding protein 4 | | Authors: | Sitar, T, Popowicz, G.M, Siwanowicz, I, Huber, R, Holak, T.A. | | Deposit date: | 2006-07-05 | | Release date: | 2006-08-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for the inhibition of insulin-like growth factors by insulin-like growth factor-binding proteins.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
2CG7
 
 | | SECOND AND THIRD FIBRONECTIN TYPE I MODULE PAIR (CRYSTAL FORM II). | | Descriptor: | FIBRONECTIN | | Authors: | Rudino-Pinera, E, Ravelli, R.B.G, Sheldrick, G.M, Nanao, M.H, Werner, J.M, Schwarz-Linek, U, Potts, J.R, Garman, E.F. | | Deposit date: | 2006-02-27 | | Release date: | 2007-02-27 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | The Solution and Crystal Structures of a Module Pair from the Staphylococcus Aureus-Binding Site of Human Fibronectin-A Tale with a Twist.
J.Mol.Biol., 368, 2007
|
|
2CBH
 
 | |
2CHD
 
 | | Crystal structure of the C2A domain of Rabphilin-3A | | Descriptor: | GLYCEROL, RABPHILIN-3A | | Authors: | Biadene, M, Montaville, P, Sheldrick, G.M, Becker, S. | | Deposit date: | 2006-03-14 | | Release date: | 2006-06-28 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Structure of the C2A Domain of Rabphilin-3A.
Acta Crystallogr.,Sect.D, 62, 2006
|
|
2CG6
 
 | | Second and third fibronectin type I module pair (crystal form I). | | Descriptor: | HUMAN FIBRONECTIN, SULFATE ION | | Authors: | Rudino-Pinera, E, Ravelli, R.B.G, Sheldrick, G.M, Nanao, M.H, Werner, J.M, Schwarz-Linek, U, Potts, J.R, Garman, E.F. | | Deposit date: | 2006-02-27 | | Release date: | 2007-02-27 | | Last modified: | 2017-08-30 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | The Solution and Crystal Structures of a Module Pair from the Staphylococcus Aureus-Binding Site of Human Fibronectin-A Tale with a Twist.
J.Mol.Biol., 368, 2007
|
|
2BBM
 
 | | SOLUTION STRUCTURE OF A CALMODULIN-TARGET PEPTIDE COMPLEX BY MULTIDIMENSIONAL NMR | | Descriptor: | CALCIUM ION, CALMODULIN, MYOSIN LIGHT CHAIN KINASE | | Authors: | Clore, G.M, Bax, A, Ikura, M, Gronenborn, A.M. | | Deposit date: | 1992-07-16 | | Release date: | 1994-01-31 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a calmodulin-target peptide complex by multidimensional NMR.
Science, 256, 1992
|
|
2BBN
 
 | | SOLUTION STRUCTURE OF A CALMODULIN-TARGET PEPTIDE COMPLEX BY MULTIDIMENSIONAL NMR | | Descriptor: | CALCIUM ION, CALMODULIN, MYOSIN LIGHT CHAIN KINASE | | Authors: | Clore, G.M, Bax, A, Ikura, M, Gronenborn, A.M. | | Deposit date: | 1992-07-16 | | Release date: | 1994-01-31 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a calmodulin-target peptide complex by multidimensional NMR.
Science, 256, 1992
|
|
2CM6
 
 | | crystal structure of the C2B domain of rabphilin3A | | Descriptor: | CALCIUM ION, PHOSPHATE ION, RABPHILIN-3A | | Authors: | Schlicker, C, Montaville, P, Sheldrick, G.M, Becker, S. | | Deposit date: | 2006-05-04 | | Release date: | 2006-12-04 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | The C2A-C2B Linker Defines the High Affinity Ca2+ Binding Mode of Rabphilin-3A.
J.Biol.Chem., 282, 2007
|
|
2EU2
 
 | | Human Carbonic Anhydrase II in complex with novel inhibitors | | Descriptor: | (R)-1-AMINO-1-[5-(DIMETHYLAMINO)-1,3,4-THIADIAZOL-2-YL]METHANESULFONAMIDE, Carbonic anhydrase 2, ZINC ION | | Authors: | Fisher, S.Z, Govindasamy, L, Boyle, N, Agbandje-McKenna, M, Silverman, D.N, Blackburn, G.M, McKenna, R. | | Deposit date: | 2005-10-28 | | Release date: | 2006-07-11 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | X-ray crystallographic studies reveal that the incorporation of spacer groups in carbonic anhydrase inhibitors causes alternate binding modes.
Acta Crystallogr.,Sect.F, 62, 2006
|
|
2EVA
 
 | | Structural Basis for the Interaction of TAK1 Kinase with its Activating Protein TAB1 | | Descriptor: | ADENOSINE, TAK1 kinase - TAB1 chimera fusion protein | | Authors: | Brown, K, Vial, S.C, Dedi, N, Long, J.M, Dunster, N.J, Cheetham, G.M. | | Deposit date: | 2005-10-31 | | Release date: | 2006-05-02 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for the interaction of TAK1 kinase with its activating protein TAB1
J.Mol.Biol., 354, 2005
|
|
2EZN
 
 | |
2EU3
 
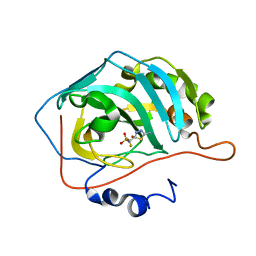 | | Human Carbonic anhydrase II in complex with novel inhibitors | | Descriptor: | 1-(5-AMINO-1,3,4-THIADIAZOL-2-YL)-1,1-DIFLUOROMETHANESULFONAMIDE, Carbonic anhydrase 2, ZINC ION | | Authors: | Fisher, S.Z, Govindasamy, L, Boyle, N, Agbandje-McKenna, M, Silverman, D.N, Blackburn, G.M, McKenna, R. | | Deposit date: | 2005-10-28 | | Release date: | 2006-07-11 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | X-ray crystallographic studies reveal that the incorporation of spacer groups in carbonic anhydrase inhibitors causes alternate binding modes.
Acta Crystallogr.,Sect.F, 62, 2006
|
|
2EZZ
 
 | |
2F3J
 
 | |
