2WF8
 
 | | Structure of Beta-Phosphoglucomutase inhibited with Glucose-6- phosphate, Glucose-1-phosphate and Beryllium trifluoride | | Descriptor: | 1-O-phosphono-alpha-D-glucopyranose, 6-O-phosphono-beta-D-glucopyranose, BERYLLIUM TRIFLUORIDE ION, ... | | Authors: | Bowler, M.W, Baxter, N.J, Webster, C.E, Pollard, S, Alizadeh, T, Hounslow, A.M, Cliff, M.J, Bermel, W, Williams, N.H, Hollfelder, F, Blackburn, G.M, Waltho, J.P. | | Deposit date: | 2009-04-03 | | Release date: | 2010-05-26 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Near attack conformers dominate beta-phosphoglucomutase complexes where geometry and charge distribution reflect those of substrate.
Proc. Natl. Acad. Sci. U.S.A., 109, 2012
|
|
2WZB
 
 | | The catalytically active fully closed conformation of human phosphoglycerate kinase in complex with ADP, 3PG and magnesium trifluoride | | Descriptor: | 3-PHOSPHOGLYCERIC ACID, ADENOSINE-5'-DIPHOSPHATE, CHLORIDE ION, ... | | Authors: | Bowler, M.W, Cliff, M.J, Marston, J.P.M, Baxter, N.J, Hownslow, A.M.H, Varga, A.V, Szabo, J, Vas, M, Blackburn, G.M, Waltho, J.P. | | Deposit date: | 2009-11-27 | | Release date: | 2010-04-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Transition State Analogue Structures of Human Phosphoglycerate Kinase Establish the Importance of Charge Balance in Catalysis.
J.Am.Chem.Soc., 132, 2010
|
|
2X13
 
 | | The catalytically active fully closed conformation of human phosphoglycerate kinase in complex with ADP and 3phosphoglycerate | | Descriptor: | 3-PHOSPHOGLYCERIC ACID, ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Bowler, M.W, Cliff, M.J, Marston, J.P.M, Baxter, N.J, Hownslow, A.M.H, Varga, A.V, Szabo, J, Vas, M, Blackburn, G.M, Waltho, J.P. | | Deposit date: | 2009-12-21 | | Release date: | 2010-12-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | The Structure of Human Phosphoglycerate Kinase in its Fully Active Conformation with Ground State Analogues
To be Published
|
|
2X15
 
 | | The catalytically active fully closed conformation of human phosphoglycerate kinase in complex with ADP and 1,3- bisphosphoglycerate | | Descriptor: | 1,3-BISPHOSPHOGLYCERIC ACID, 3-PHOSPHOGLYCERIC ACID, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Bowler, M.W, Cliff, M.J, Marston, J.P.M, Baxter, N.J, Hounslow, A.M.H, Varga, A.V, Szabo, J, Vas, M, Blackburn, G.M, Waltho, J.P. | | Deposit date: | 2009-12-21 | | Release date: | 2011-02-09 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The Structure of Human Phosphoglycerate Kinase in its Fully Active Conformation in Complex with Ground State Analoges
To be Published
|
|
2WZC
 
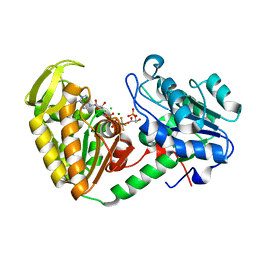 | | The catalytically active fully closed conformation of human phosphoglycerate kinase in complex with ADP, 3PG and aluminium tetrafluoride | | Descriptor: | 3-PHOSPHOGLYCERIC ACID, ADENOSINE-5'-DIPHOSPHATE, CHLORIDE ION, ... | | Authors: | Bowler, M.W, Cliff, M.J, Marston, J.P.M, Baxter, N.J, Hownslow, A.M.H, Varga, A.V, Szabo, J, Vas, M, Blackburn, G.M, Waltho, J.P. | | Deposit date: | 2009-11-27 | | Release date: | 2010-04-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Transition State Analogue Structures of Human Phosphoglycerate Kinase Establish the Importance of Charge Balance in Catalysis.
J.Am.Chem.Soc., 132, 2010
|
|
2X14
 
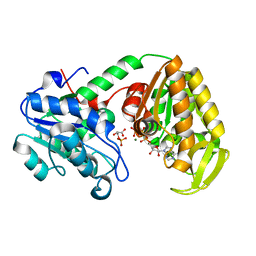 | | The catalytically active fully closed conformation of human phosphoglycerate kinase K219A mutant in complex with AMP-PCP and 3PG | | Descriptor: | 3-PHOSPHOGLYCERIC ACID, MAGNESIUM ION, PHOSPHOGLYCERATE KINASE 1, ... | | Authors: | Bowler, M.W, Cliff, M.J, Marston, J.P.M, Baxter, N.J, Hownslow, A.M.H, Varga, A.V, Szabo, J, Vas, M, Blackburn, G.M, Waltho, J.P. | | Deposit date: | 2009-12-21 | | Release date: | 2010-12-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The Structure of Human Phosphoglycerate Kinase in its Fully Active Conformation in Complex with Ground State Analogues
To be Published
|
|
2WFA
 
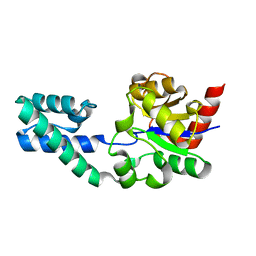 | | Structure of Beta-Phosphoglucomutase inhibited with Beryllium trifluoride, in an open conformation. | | Descriptor: | BERYLLIUM TRIFLUORIDE ION, BETA-PHOSPHOGLUCOMUTASE, MAGNESIUM ION | | Authors: | Bowler, M.W, Baxter, N.J, Webster, C.E, Pollard, S, Alizadeh, T, Hounslow, A.M, Cliff, M.J, Bermel, W, Williams, N.H, Hollfelder, F, Blackburn, G.M, Waltho, J.P. | | Deposit date: | 2009-04-03 | | Release date: | 2010-05-26 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Near Attack Conformers Dominate Beta-Phosphoglucomutase Complexes Where Geometry and Charge Distribution Reflect Those of Substrate.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
2XDF
 
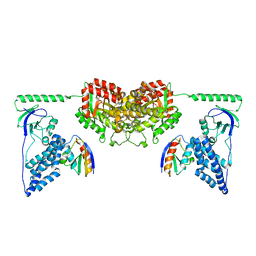 | | Solution Structure of the Enzyme I Dimer Complexed with HPr Using Residual Dipolar Couplings and Small Angle X-Ray Scattering | | Descriptor: | PHOSPHOCARRIER PROTEIN HPR, PHOSPHOENOLPYRUVATE-PROTEIN PHOSPHOTRANSFERASE | | Authors: | Schwieters, C.D, Suh, J.-Y, Grishaev, A, Guirlando, R, Takayama, Y, Clore, G.M. | | Deposit date: | 2010-04-30 | | Release date: | 2010-09-22 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR, SOLUTION SCATTERING | | Cite: | Solution Structure of the 128 kDa Enzyme I Dimer from Escherichia Coli and its 146 kDa Complex with Hpr Using Residual Dipolar Couplings and Small- and Wide-Angle X-Ray Scattering.
J.Am.Chem.Soc., 132, 2010
|
|
2Y8N
 
 | | Crystal structure of glycyl radical enzyme | | Descriptor: | 4-HYDROXYPHENYLACETATE DECARBOXYLASE LARGE SUBUNIT, 4-HYDROXYPHENYLACETATE DECARBOXYLASE SMALL SUBUNIT, IRON/SULFUR CLUSTER | | Authors: | Martins, B.M, Blaser, M, Feliks, M, Ullmann, G.M, Selmer, T. | | Deposit date: | 2011-02-08 | | Release date: | 2011-09-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural Basis for a Kolbe-Type Decarboxylation Catalyzed by a Glycyl Radical Enzyme.
J.Am.Chem.Soc., 133, 2011
|
|
1U12
 
 | | M. loti cyclic nucleotide binding domain mutant | | Descriptor: | IODIDE ION, POTASSIUM ION, SULFATE ION, ... | | Authors: | Clayton, G.M, Silverman, W.R, Heginbotham, L, Morais-Cabral, J.H. | | Deposit date: | 2004-07-14 | | Release date: | 2004-11-30 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural Basis of Ligand Activation in a Cyclic Nucleotide Regulated Potassium Channel
Cell(Cambridge,Mass.), 119, 2004
|
|
1RZY
 
 | | Crystal structure of rabbit Hint complexed with N-ethylsulfamoyladenosine | | Descriptor: | 5'-O-(N-ETHYL-SULFAMOYL)ADENOSINE, Histidine triad nucleotide-binding protein 1 | | Authors: | Krakowiak, A.K, Pace, H.C, Blackburn, G.M, Adams, M, Mekhalfia, A, Kaczmarek, R, Baraniak, J, Stec, W.J, Brenner, C. | | Deposit date: | 2003-12-29 | | Release date: | 2004-03-09 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Biochemical, crystallographic, and mutagenic characterization of hint, the AMP-lysine hydrolase, with novel substrates and inhibitors
J.Biol.Chem., 279, 2004
|
|
1UNJ
 
 | | Crystal structure of a 7-Aminoactinomycin D complex with non-complementary DNA | | Descriptor: | 5'-D(*TP*TP*AP*GP*BRU*TP)-3', 7-AMINO-ACTINOMYCIN D | | Authors: | Alexopoulos, E.C, Klement, R, Jares-Erijman, E.A, Uson, I, Jovin, T.M, Sheldrick, G.M. | | Deposit date: | 2003-09-10 | | Release date: | 2004-12-16 | | Last modified: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal and Solution Structures of 7-Amino-Actinomycin D Complexes with D(Ttagbrut), D(Ttagtt) and D(Tttagttt)
Acta Crystallogr.,Sect.D, 61, 2005
|
|
1RYJ
 
 | | Solution NMR Structure of Protein Mth1743 from Methanobacterium thermoautotrophicum. Ontario Centre for Structural Proteomics target MTH1743_1_70; Northeast Structural Genomics Consortium Target TT526. | | Descriptor: | unknown | | Authors: | Yee, A, Chang, X, Pineda-Lucena, A, Wu, B, Semesi, A, Le, B, Ramelot, T, Lee, G.M, Bhattacharyya, S, Gutierrez, P, Denisov, A, Lee, C.H, Cort, J.R, Kozlov, G, Liao, J, Finak, G, Chen, L, Wishart, D, Lee, W, McIntosh, L.P, Gehring, K, Kennedy, M.A, Edwards, A.M, Arrowsmith, C.H, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2003-12-22 | | Release date: | 2004-02-24 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | AN NMR APPROACH TO STRUCTURAL PROTEOMICS
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
1SHO
 
 | |
1UNM
 
 | | Crystal structure of 7-Aminoactinomycin D with non-complementary DNA | | Descriptor: | 5'-D(*TP*TP*AP*GP*BRU*TP)-3', 7-AMINOACTINOMYCIN D | | Authors: | Alexopoulos, E.C, Klement, R, Jares-Erijman, E.A, Uson, I, Jovin, T.M, Sheldrick, G.M. | | Deposit date: | 2003-09-11 | | Release date: | 2004-09-24 | | Last modified: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal and Solution Structures of 7-Amino-Actinomycin D Complexes with D(Ttagbrut), D(Ttagtt) and D(Tttagttt)
Acta Crystallogr.,Sect.D, 61, 2005
|
|
1UNO
 
 | | Crystal structure of a d,l-alternating peptide | | Descriptor: | H-(L-TYR-D-TYR)4-LYS-OH | | Authors: | Alexopoulos, E, Kuesel, A, Uson, I, Diederichsen, U, Sheldrick, G.M. | | Deposit date: | 2003-09-11 | | Release date: | 2004-09-24 | | Last modified: | 2019-07-24 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Solution and Structure of an Alternating D,L-Peptide
Acta Crystallogr.,Sect.D, 60, 2004
|
|
1VCJ
 
 | | Influenza B virus neuraminidase complexed with 1-(4-Carboxy-2-(3-pentylamino)phenyl)-5-aminomethyl-5-hydroxymethyl-pyrrolidin-2-one | | Descriptor: | 4-[(2R)-2-(AMINOMETHYL)-2-(HYDROXYMETHYL)-5-OXOPYRROLIDIN-1-YL]-3-[(1-ETHYLPROPYL)AMINO]BENZOIC ACID, NEURAMINIDASE | | Authors: | Lommer, B.S, Ali, S.M, Bajpai, S.N, Brouillette, W.J, Air, G.M, Luo, M. | | Deposit date: | 2004-03-09 | | Release date: | 2004-03-23 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | A benzoic acid inhibitor induces a novel conformational change in the active site of Influenza B virus neuraminidase.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
1VKR
 
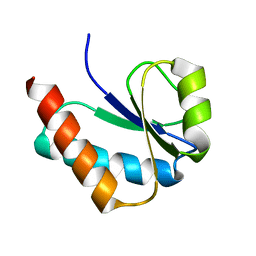 | |
1VMC
 
 | | STROMA CELL-DERIVED FACTOR-1ALPHA (SDF-1ALPHA) | | Descriptor: | Stromal cell-derived factor 1 | | Authors: | Gozansky, E.K, Clore, G.M. | | Deposit date: | 2004-09-20 | | Release date: | 2005-03-01 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Mapping the binding of the N-terminal extracellular tail of the CXCR4 receptor to stromal cell-derived factor-1alpha.
J.Mol.Biol., 345, 2005
|
|
1VP6
 
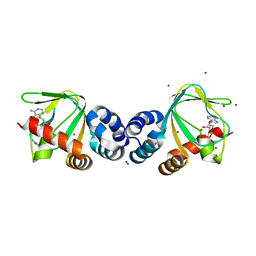 | | M.loti ion channel cylic nucleotide binding domain | | Descriptor: | ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE, BROMIDE ION, Cyclic-nucleotide binding domain of mesorhizobium loti CNG potassium channel | | Authors: | Clayton, G.M, Silverman, W.R, Heginbotham, L, Morais-Cabral, J.H. | | Deposit date: | 2004-10-14 | | Release date: | 2004-10-26 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural basis of ligand activation in a cyclic nucleotide regulated potassium channel.
Cell(Cambridge,Mass.), 119, 2004
|
|
1W7Q
 
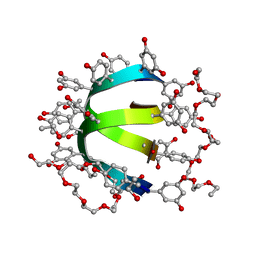 | | Feglymycin P65 crystal form | | Descriptor: | 1,2-ETHANEDIOL, ETHANOL, FEGLYMYCIN | | Authors: | Bunkoczi, G, Vertesy, L, Sheldrick, G.M. | | Deposit date: | 2004-09-08 | | Release date: | 2005-03-08 | | Last modified: | 2019-05-22 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | The Antiviral Antibiotic Feglymycin: First Direct Methods Solution of a 1000Plus Equal-Atom Structure
Angew.Chem.Int.Ed.Engl., 44, 2005
|
|
1W7R
 
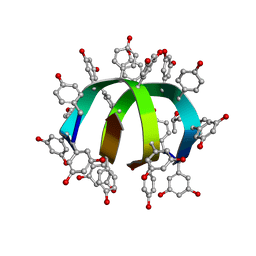 | | Feglymycin P64 crystal form | | Descriptor: | CITRATE ANION, FEGLYMYCIN, ISOPROPYL ALCOHOL | | Authors: | Bunkoczi, G, Vertesy, L, Sheldrick, G.M. | | Deposit date: | 2004-09-08 | | Release date: | 2005-03-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | The Antiviral Antibiotic Feglymycin: First Direct Methods Solution of a 1000Plus Equal-Atom Structure
Angew.Chem.Int.Ed.Engl., 44, 2005
|
|
1VRC
 
 | |
1VRV
 
 | |
1WJB
 
 | | SOLUTION STRUCTURE OF THE N-TERMINAL ZN BINDING DOMAIN OF HIV-1 INTEGRASE (D FORM), NMR, 40 STRUCTURES | | Descriptor: | HIV-1 INTEGRASE, ZINC ION | | Authors: | Clore, G.M, Cai, M, Caffrey, M, Gronenborn, A.M. | | Deposit date: | 1997-05-13 | | Release date: | 1998-05-13 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the N-terminal zinc binding domain of HIV-1 integrase.
Nat.Struct.Biol., 4, 1997
|
|
