6BAA
 
 | | Cryo-EM structure of the pancreatic beta-cell KATP channel bound to ATP and glibenclamide | | 分子名称: | 5-chloro-N-(2-{4-[(cyclohexylcarbamoyl)sulfamoyl]phenyl}ethyl)-2-methoxybenzamide, ADENOSINE-5'-TRIPHOSPHATE, ATP-binding cassette sub-family C member 8, ... | | 著者 | Martin, G.M, Yoshioka, C, Shyng, S.L. | | 登録日 | 2017-10-12 | | 公開日 | 2017-11-01 | | 最終更新日 | 2019-12-25 | | 実験手法 | ELECTRON MICROSCOPY (3.63 Å) | | 主引用文献 | Anti-diabetic drug binding site in a mammalian KATPchannel revealed by Cryo-EM.
Elife, 6, 2017
|
|
6B9C
 
 | | Superfolder Green Fluorescent Protein with 4-nitro-L-phenylalanine at the chromophore (position 66) | | 分子名称: | CARBON DIOXIDE, Green fluorescent protein | | 著者 | Phillips-Piro, C.M, Brewer, S.H, Olenginski, G.M, Piacentini, J. | | 登録日 | 2017-10-10 | | 公開日 | 2018-10-17 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (1.695 Å) | | 主引用文献 | Structural and spectrophotometric investigation of two unnatural amino-acid altered chromophores in the superfolder green fluorescent protein
Acta Crystallogr.,Sect.D, 2021
|
|
6ARQ
 
 | | Crystal structure of CD96 (D1) bound to CD155/necl-5 (D1-3) | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Poliovirus receptor, T-cell surface protein tactile, ... | | 著者 | Deuss, F.A, Watson, G.M, Rossjohn, J, Berry, R. | | 登録日 | 2017-08-23 | | 公開日 | 2018-11-21 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.88 Å) | | 主引用文献 | Structural Basis for CD96 Immune Receptor Recognition of Nectin-like Protein-5, CD155.
Structure, 27, 2019
|
|
6BDO
 
 | |
6C44
 
 | | Zika virus capsid protein | | 分子名称: | Capsid protein | | 著者 | Morando, M.A, Barbosa, G.M, Cruz-Oliveira, C, Da Poian, A.T, Almeida, F.C.L. | | 登録日 | 2018-01-11 | | 公開日 | 2019-01-16 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Dynamics of Zika Virus Capsid Protein in Solution: The Properties and Exposure of the Hydrophobic Cleft Are Controlled by the alpha-Helix 1 Sequence.
Biochemistry, 58, 2019
|
|
6BJX
 
 | |
6D8M
 
 | |
6D8O
 
 | |
7YK1
 
 | | Structural basis of human PRPS2 filaments | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, PHOSPHATE ION, ... | | 著者 | Lu, G.M, Hu, H.H, Liu, J.L. | | 登録日 | 2022-07-21 | | 公開日 | 2023-08-02 | | 最終更新日 | 2023-09-20 | | 実験手法 | ELECTRON MICROSCOPY (3.08 Å) | | 主引用文献 | Structural basis of human PRPS2 filaments.
Cell Biosci, 13, 2023
|
|
7YLT
 
 | |
7YLS
 
 | | Structure of a bacteria protein complex | | 分子名称: | ACETATE ION, Aromatic-ring-hydroxylating dioxygenase beta subunit, FE (III) ION, ... | | 著者 | Zhang, H, Ma, Y.J, Yu, G.M, Li, X.Z. | | 登録日 | 2022-07-26 | | 公開日 | 2023-06-14 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Structure of a bacteria protein complex
To Be Published
|
|
7YLR
 
 | |
7XRV
 
 | |
7XRT
 
 | |
6N8C
 
 | | Structure of the Huntingtin tetramer/dimer mixture determined by paramagnetic NMR | | 分子名称: | Huntingtin | | 著者 | Schwieters, C.D, Kotler, S.A, Schmidt, T, Ceccon, A, Ghirlando, R, Libich, D.S, Clore, G.M. | | 登録日 | 2018-11-29 | | 公開日 | 2019-02-13 | | 最終更新日 | 2024-05-01 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Probing initial transient oligomerization events facilitating Huntingtin fibril nucleation at atomic resolution by relaxation-based NMR.
Proc. Natl. Acad. Sci. U.S.A., 116, 2019
|
|
6XIR
 
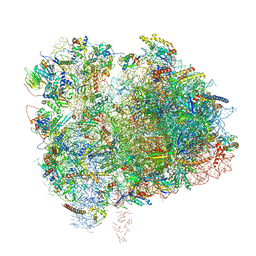 | | Cryo-EM Structure of K63 Ubiquitinated Yeast Translocating Ribosome under Oxidative Stress | | 分子名称: | 18S ribosomal RNA, 35S ribosomal RNA, 40S ribosomal protein S0-A, ... | | 著者 | Zhou, Y, Bartesaghi, A, Silva, G.M. | | 登録日 | 2020-06-21 | | 公開日 | 2020-08-26 | | 最終更新日 | 2020-09-23 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | Structural impact of K63 ubiquitin on yeast translocating ribosomes under oxidative stress.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6YI4
 
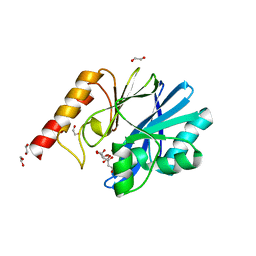 | | Structure of IMP-13 metallo-beta-lactamase complexed with citrate anion | | 分子名称: | 1,2-ETHANEDIOL, ACETATE ION, BETA-MERCAPTOETHANOL, ... | | 著者 | Zak, K.M, Zhou, R.X, Softley, C.A, Bostock, M.J, Sattler, M, Popowicz, G.M. | | 登録日 | 2020-03-31 | | 公開日 | 2020-04-08 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Structure of IMP-13 metallo-beta-lactamase complexed with citrate anion
Not published
|
|
6XIQ
 
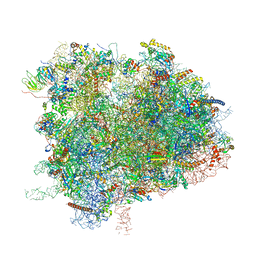 | | Cryo-EM Structure of K63R Ubiquitin Mutant Ribosome under Oxidative Stress | | 分子名称: | 18S ribosomal RNA, 35S ribosomal RNA, 40S ribosomal protein S0-A, ... | | 著者 | Zhou, Y, Bartesaghi, A, Silva, G.M. | | 登録日 | 2020-06-21 | | 公開日 | 2020-08-26 | | 最終更新日 | 2020-09-23 | | 実験手法 | ELECTRON MICROSCOPY (4.2 Å) | | 主引用文献 | Structural impact of K63 ubiquitin on yeast translocating ribosomes under oxidative stress.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
3GAT
 
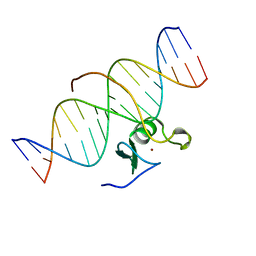 | | SOLUTION NMR STRUCTURE OF THE C-TERMINAL DOMAIN OF CHICKEN GATA-1 BOUND TO DNA, 34 STRUCTURES | | 分子名称: | DNA (5'-D(*AP*AP*TP*GP*TP*TP*TP*AP*TP*CP*TP*GP*CP*AP*AP*C)-3'), DNA (5'-D(*GP*TP*TP*GP*CP*AP*GP*AP*TP*AP*AP*AP*CP*AP*TP*T)-3'), ERYTHROID TRANSCRIPTION FACTOR GATA-1, ... | | 著者 | Clore, G.M, Tjandra, N, Starich, M, Omichinski, J.G, Gronenborn, A.M. | | 登録日 | 1997-11-07 | | 公開日 | 1998-01-28 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Use of dipolar 1H-15N and 1H-13C couplings in the structure determination of magnetically oriented macromolecules in solution.
Nat.Struct.Biol., 4, 1997
|
|
3GT4
 
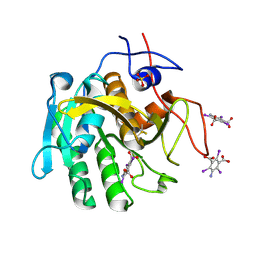 | | Structure of proteinase K with the magic triangle I3C | | 分子名称: | 5-amino-2,4,6-triiodobenzene-1,3-dicarboxylic acid, SULFATE ION, proteinase K | | 著者 | Beck, T, Gruene, T, Sheldrick, G.M. | | 登録日 | 2009-03-27 | | 公開日 | 2009-04-14 | | 最終更新日 | 2017-11-01 | | 実験手法 | X-RAY DIFFRACTION (1.76 Å) | | 主引用文献 | The magic triangle goes MAD: experimental phasing with a bromine derivative
Acta Crystallogr.,Sect.D, 66, 2010
|
|
3GT3
 
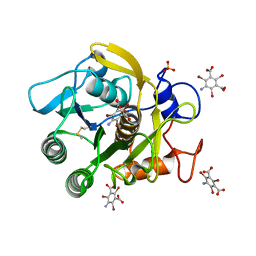 | | Structure of proteinase K with the mad triangle B3C | | 分子名称: | 5-amino-2,4,6-tribromobenzene-1,3-dicarboxylic acid, Proteinase K, SULFATE ION | | 著者 | Beck, T, Gruene, T, Sheldrick, G.M. | | 登録日 | 2009-03-27 | | 公開日 | 2009-04-14 | | 最終更新日 | 2017-11-01 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | The magic triangle goes MAD: experimental phasing with a bromine derivative
Acta Crystallogr.,Sect.D, 66, 2010
|
|
5ADO
 
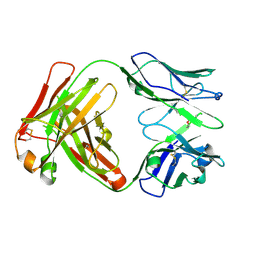 | | Crystal structure of the paraoxon-modified A.17 antibody FAB fragment - Light chain S35R mutant | | 分子名称: | DIETHYL PHOSPHONATE, FAB A.17 | | 著者 | Chatziefthimiou, S.D, Smirnov, I.V, Golovin, A.V, Stepanova, A.V, Peng, Y, Zolotareva, O.I, Belogurov, A.A, Ponomarenko, N.A, Blackburn, G.M, Gabibov, A.A, Lerner, R, Wilmanns, M. | | 登録日 | 2015-08-21 | | 公開日 | 2016-11-09 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.55 Å) | | 主引用文献 | Robotic Qm/Mm-Driven Maturation of Antibody Combining Sites.
Sci.Adv., 2, 2016
|
|
6O3O
 
 | | Structure of human DNAM-1 (CD226) bound to nectin-like protein-5 (necl-5) | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CD226 antigen, ... | | 著者 | Deuss, F.A, Watson, G.M, Rossjohn, J, Berry, R. | | 登録日 | 2019-02-27 | | 公開日 | 2019-07-10 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Structural basis for the recognition of nectin-like protein-5 by the human-activating immune receptor, DNAM-1.
J.Biol.Chem., 294, 2019
|
|
3ITI
 
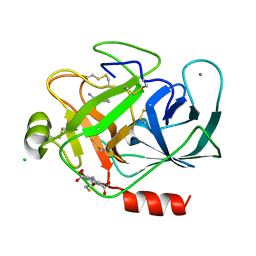 | | Structure of bovine trypsin with the MAD triangle B3C | | 分子名称: | 5-amino-2,4,6-tribromobenzene-1,3-dicarboxylic acid, BENZAMIDINE, CALCIUM ION, ... | | 著者 | Beck, T, da Cunha, C.E, Sheldrick, G.M. | | 登録日 | 2009-08-28 | | 公開日 | 2009-10-27 | | 最終更新日 | 2018-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.55 Å) | | 主引用文献 | How to get the magic triangle and the MAD triangle into your protein crystal.
Acta Crystallogr.,Sect.F, 65, 2009
|
|
7SJR
 
 | | Cryo-EM structure of AdnA-AdnB(W325A) in complex with DNA and AMPPNP | | 分子名称: | DNA (70-MER), DNA helicase, IRON/SULFUR CLUSTER, ... | | 著者 | Wang, J, Warren, G.M, Shuman, S, Patel, D.J. | | 登録日 | 2021-10-18 | | 公開日 | 2021-12-22 | | 最終更新日 | 2024-06-05 | | 実験手法 | ELECTRON MICROSCOPY (3.8 Å) | | 主引用文献 | Structure-activity relationships at a nucleobase-stacking tryptophan required for chemomechanical coupling in the DNA resecting motor-nuclease AdnAB.
Nucleic Acids Res., 50, 2022
|
|
