1AEV
 
 | | INTRODUCTION OF NOVEL SUBSTRATE OXIDATION INTO CYTOCHROME C PEROXIDASE BY CAVITY COMPLEMENTATION: OXIDATION OF 2-AMINOTHIAZOLE AND COVALENT MODIFICATION OF THE ENZYME (2-AMINOTHIAZOLE) | | 分子名称: | 2-AMINOTHIAZOLE, CYTOCHROME C PEROXIDASE, PROTOPORPHYRIN IX CONTAINING FE | | 著者 | Musah, R.A, Fitzgerald, M.M, Jensen, G.M, Mcree, D.E, Goodin, D.B. | | 登録日 | 1997-02-25 | | 公開日 | 1997-09-04 | | 最終更新日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Introduction of novel substrate oxidation into cytochrome c peroxidase by cavity complementation: oxidation of 2-aminothiazole and covalent modification of the enzyme.
Biochemistry, 36, 1997
|
|
1EMV
 
 | | CRYSTAL STRUCTURE OF COLICIN E9 DNASE DOMAIN WITH ITS COGNATE IMMUNITY PROTEIN IM9 (1.7 ANGSTROMS) | | 分子名称: | COLICIN E9, IMMUNITY PROTEIN IM9, PHOSPHATE ION | | 著者 | Kuhlmann, U.C, Pommer, A.J, Moore, G.M, James, R, Kleanthous, C. | | 登録日 | 2000-03-20 | | 公開日 | 2000-09-13 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Specificity in protein-protein interactions: the structural basis for dual recognition in endonuclease colicin-immunity protein complexes.
J.Mol.Biol., 301, 2000
|
|
7JG7
 
 | | Cryo-EM structure of bedaquiline-free Mycobacterium smegmatis ATP synthase rotational state 3 (backbone model) | | 分子名称: | ATP synthase epsilon chain, ATP synthase gamma chain, ATP synthase subunit a, ... | | 著者 | Guo, H, Courbon, G.M, Rubinstein, J.L. | | 登録日 | 2020-07-18 | | 公開日 | 2020-08-19 | | 最終更新日 | 2024-03-06 | | 実験手法 | ELECTRON MICROSCOPY (3.5 Å) | | 主引用文献 | Structure of mycobacterial ATP synthase bound to the tuberculosis drug bedaquiline.
Nature, 589, 2021
|
|
7JG8
 
 | | Cryo-EM structure of bedaquiline-saturated Mycobacterium smegmatis ATP synthase rotational state 1 (backbone model) | | 分子名称: | ATP synthase epsilon chain, ATP synthase gamma chain, ATP synthase subunit a, ... | | 著者 | Guo, H, Courbon, G.M, Rubinstein, J.L. | | 登録日 | 2020-07-18 | | 公開日 | 2020-08-19 | | 最終更新日 | 2024-03-06 | | 実験手法 | ELECTRON MICROSCOPY (3.3 Å) | | 主引用文献 | Structure of mycobacterial ATP synthase bound to the tuberculosis drug bedaquiline.
Nature, 589, 2021
|
|
7JGB
 
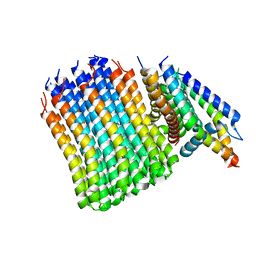 | | Cryo-EM structure of bedaquiline-free Mycobacterium smegmatis ATP synthase FO region | | 分子名称: | ATP synthase subunit a, ATP synthase subunit b, ATP synthase subunit b-delta, ... | | 著者 | Guo, H, Courbon, G.M, Rubinstein, J.L. | | 登録日 | 2020-07-18 | | 公開日 | 2020-08-19 | | 最終更新日 | 2024-03-06 | | 実験手法 | ELECTRON MICROSCOPY (3.5 Å) | | 主引用文献 | Structure of mycobacterial ATP synthase bound to the tuberculosis drug bedaquiline.
Nature, 589, 2021
|
|
7JGA
 
 | | Cryo-EM structure of bedaquiline-saturated Mycobacterium smegmatis ATP synthase rotational state 3 | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP synthase epsilon chain, ... | | 著者 | Guo, H, Courbon, G.M, Rubinstein, J.L. | | 登録日 | 2020-07-18 | | 公開日 | 2020-08-19 | | 最終更新日 | 2024-03-06 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | Structure of mycobacterial ATP synthase bound to the tuberculosis drug bedaquiline.
Nature, 589, 2021
|
|
7JG6
 
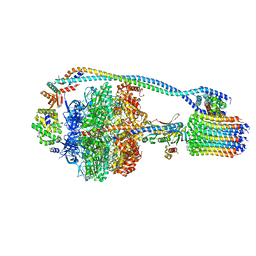 | | Cryo-EM structure of bedaquiline-free Mycobacterium smegmatis ATP synthase rotational state 2 (backbone model) | | 分子名称: | ATP synthase epsilon chain, ATP synthase gamma chain, ATP synthase subunit a, ... | | 著者 | Guo, H, Courbon, G.M, Rubinstein, J.L. | | 登録日 | 2020-07-18 | | 公開日 | 2020-08-19 | | 最終更新日 | 2024-03-06 | | 実験手法 | ELECTRON MICROSCOPY (3.7 Å) | | 主引用文献 | Structure of mycobacterial ATP synthase bound to the tuberculosis drug bedaquiline.
Nature, 589, 2021
|
|
7JG5
 
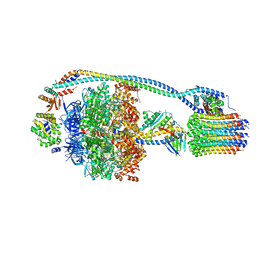 | | Cryo-EM structure of bedaquiline-free Mycobacterium smegmatis ATP synthase rotational state 1 | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP synthase epsilon chain, ... | | 著者 | Guo, H, Courbon, G.M, Rubinstein, J.L. | | 登録日 | 2020-07-18 | | 公開日 | 2020-08-19 | | 最終更新日 | 2024-03-06 | | 実験手法 | ELECTRON MICROSCOPY (3.4 Å) | | 主引用文献 | Structure of mycobacterial ATP synthase bound to the tuberculosis drug bedaquiline.
Nature, 589, 2021
|
|
7JGC
 
 | | Cryo-EM structure of bedaquiline-saturated Mycobacterium smegmatis ATP synthase FO region | | 分子名称: | ATP synthase subunit a, ATP synthase subunit b, ATP synthase subunit b-delta, ... | | 著者 | Guo, H, Courbon, G.M, Rubinstein, J.L. | | 登録日 | 2020-07-18 | | 公開日 | 2020-08-19 | | 最終更新日 | 2024-03-06 | | 実験手法 | ELECTRON MICROSCOPY (3.4 Å) | | 主引用文献 | Structure of mycobacterial ATP synthase bound to the tuberculosis drug bedaquiline.
Nature, 589, 2021
|
|
7JG9
 
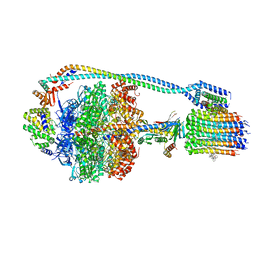 | | Cryo-EM structure of bedaquiline-saturated mycobacterium smegmatis ATP synthase rotational state 2 (backbone model) | | 分子名称: | ATP synthase epsilon chain, ATP synthase gamma chain, ATP synthase subunit a, ... | | 著者 | Guo, H, Courbon, G.M, Rubinstein, J.L. | | 登録日 | 2020-07-18 | | 公開日 | 2020-08-19 | | 最終更新日 | 2024-03-06 | | 実験手法 | ELECTRON MICROSCOPY (3.4 Å) | | 主引用文献 | Structure of mycobacterial ATP synthase bound to the tuberculosis drug bedaquiline.
Nature, 589, 2021
|
|
7GAT
 
 | | SOLUTION NMR STRUCTURE OF THE L22V MUTANT DNA BINDING DOMAIN OF AREA COMPLEXED TO A 13 BP DNA CONTAINING A TGATA SITE, 34 STRUCTURES | | 分子名称: | DNA (5'-D(*CP*AP*GP*TP*GP*AP*TP*AP*GP*AP*GP*AP*C)-3'), DNA (5'-D(*GP*TP*CP*TP*CP*TP*AP*TP*CP*AP*CP*TP*G)-3'), NITROGEN REGULATORY PROTEIN AREA, ... | | 著者 | Clore, G.M, Starich, M, Wikstrom, M, Gronenborn, A.M. | | 登録日 | 1997-11-07 | | 公開日 | 1998-01-28 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | The solution structure of the Leu22-->Val mutant AREA DNA binding domain complexed with a TGATAG core element defines a role for hydrophobic packing in the determination of specificity.
J.Mol.Biol., 277, 1998
|
|
5RHD
 
 | | PanDDA analysis group deposition SARS-CoV-2 main protease fragment screen -- Crystal Structure of SARS-CoV-2 main protease in complex with SF013 (Mpro-x2193) | | 分子名称: | 1-[4-(methylsulfonyl)phenyl]piperazine, 3C-like proteinase, DIMETHYL SULFOXIDE | | 著者 | Fearon, D, Owen, C.D, Douangamath, A, Lukacik, P, Powell, A.J, Strain-Damerell, C.M, Resnick, E, Krojer, T, Gehrtz, P, Wild, C, Aimon, A, Brandao-Neto, J, Carbery, A, Dunnett, L, Skyner, R, Snee, M, Keeley, A, Keseru, G.M, London, N, Walsh, M.A, von Delft, F. | | 登録日 | 2020-05-16 | | 公開日 | 2020-06-10 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (1.57 Å) | | 主引用文献 | Crystallographic and electrophilic fragment screening of the SARS-CoV-2 main protease.
Nat Commun, 11, 2020
|
|
5O5Q
 
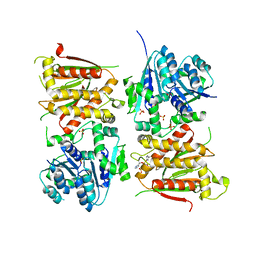 | | X-ray crystal structure of RapZ from Escherichia coli (P3221 space group) | | 分子名称: | RNase adapter protein RapZ, SULFATE ION | | 著者 | Gonzalez, G.M, Durica-Mitic, S, Hardwick, S.W, Moncrieffe, M, Resch, M, Neumann, P, Ficner, R, Gorke, B, Luisi, B.F. | | 登録日 | 2017-06-02 | | 公開日 | 2017-08-30 | | 最終更新日 | 2017-11-01 | | 実験手法 | X-RAY DIFFRACTION (3.25 Å) | | 主引用文献 | Structural insights into RapZ-mediated regulation of bacterial amino-sugar metabolism.
Nucleic Acids Res., 45, 2017
|
|
5O5O
 
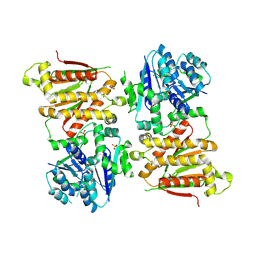 | | X-ray crystal structure of RapZ from Escherichia coli (P32 space group) | | 分子名称: | RNase adapter protein RapZ, SULFATE ION | | 著者 | Gonzalez, G.M, Durica-Mitic, S, Hardwick, S.W, Moncrieffe, M, Resch, M, Neumann, P, Ficner, R, Gorke, B, Luisi, B.F. | | 登録日 | 2017-06-02 | | 公開日 | 2017-08-30 | | 最終更新日 | 2017-11-01 | | 実験手法 | X-RAY DIFFRACTION (3.404 Å) | | 主引用文献 | Structural insights into RapZ-mediated regulation of bacterial amino-sugar metabolism.
Nucleic Acids Res., 45, 2017
|
|
5RHC
 
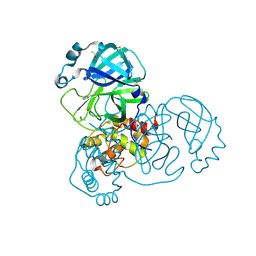 | | PanDDA analysis group deposition SARS-CoV-2 main protease fragment screen -- Crystal Structure of SARS-CoV-2 main protease in complex with Cov_HetLib053 (Mpro-x2119) | | 分子名称: | (E)-1-(1H-imidazol-2-yl)methanimine, 3C-like proteinase, DIMETHYL SULFOXIDE | | 著者 | Fearon, D, Owen, C.D, Douangamath, A, Lukacik, P, Powell, A.J, Strain-Damerell, C.M, Resnick, E, Krojer, T, Gehrtz, P, Wild, C, Aimon, A, Brandao-Neto, J, Carbery, A, Dunnett, L, Skyner, R, Snee, M, Keeley, A, Keseru, G.M, London, N, Walsh, M.A, von Delft, F. | | 登録日 | 2020-05-16 | | 公開日 | 2020-06-10 | | 最終更新日 | 2021-02-24 | | 実験手法 | X-RAY DIFFRACTION (1.58 Å) | | 主引用文献 | Crystallographic and electrophilic fragment screening of the SARS-CoV-2 main protease.
Nat Commun, 11, 2020
|
|
5RHB
 
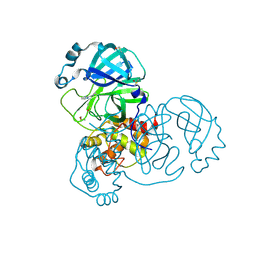 | | PanDDA analysis group deposition SARS-CoV-2 main protease fragment screen -- Crystal Structure of SARS-CoV-2 main protease in complex with Cov_HetLib030 (Mpro-x2097) | | 分子名称: | (E)-1-(pyrimidin-2-yl)methanimine, 3C-like proteinase, DIMETHYL SULFOXIDE | | 著者 | Fearon, D, Owen, C.D, Douangamath, A, Lukacik, P, Powell, A.J, Strain-Damerell, C.M, Resnick, E, Krojer, T, Gehrtz, P, Wild, C, Aimon, A, Brandao-Neto, J, Carbery, A, Dunnett, L, Skyner, R, Snee, M, Keeley, A, Keseru, G.M, London, N, Walsh, M.A, von Delft, F. | | 登録日 | 2020-05-16 | | 公開日 | 2020-06-10 | | 最終更新日 | 2021-02-24 | | 実験手法 | X-RAY DIFFRACTION (1.43 Å) | | 主引用文献 | Crystallographic and electrophilic fragment screening of the SARS-CoV-2 main protease.
Nat Commun, 11, 2020
|
|
5S71
 
 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 NendoU in complex with FUZS-5 | | 分子名称: | 5'-thiothymidine, CITRIC ACID, Uridylate-specific endoribonuclease | | 著者 | Godoy, A.S, Bege, M, Bajusz, D, BorbAs, A, Keseru, G.M, Douangamath, A, Nakamura, A.M, Dias, A, Krojer, T, Noske, G.D, Gawiljuk, V.O, Fernandes, R.S, Fairhead, M, Powell, A, Dunnet, L, Aimon, A, Fearon, D, Brandao-Neto, J, Skyner, R, von Delft, F, Oliva, G. | | 登録日 | 2020-11-13 | | 公開日 | 2020-11-25 | | 最終更新日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (1.941 Å) | | 主引用文献 | Allosteric regulation and crystallographic fragment screening of SARS-CoV-2 NSP15 endoribonuclease.
Nucleic Acids Res., 2023
|
|
7DJJ
 
 | | Structure of four truncated and mutated forms of quenching protein lumenal domains | | 分子名称: | Protein SUPPRESSOR OF QUENCHING 1, chloroplastic, SODIUM ION, ... | | 著者 | Yu, G.M, Pan, X.W, Li, M. | | 登録日 | 2020-11-20 | | 公開日 | 2022-06-08 | | 最終更新日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (2.69806433 Å) | | 主引用文献 | Structure of Arabidopsis SOQ1 lumenal region unveils C-terminal domain essential for negative regulation of photoprotective qH.
Nat.Plants, 8, 2022
|
|
7DJM
 
 | | Structure of four truncated and mutated forms of quenching protein | | 分子名称: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, ACETATE ION, Protein SUPPRESSOR OF QUENCHING 1, ... | | 著者 | Yu, G.M, Pan, X.W, Li, M. | | 登録日 | 2020-11-20 | | 公開日 | 2022-06-08 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.70000112 Å) | | 主引用文献 | Structure of Arabidopsis SOQ1 lumenal region unveils C-terminal domain essential for negative regulation of photoprotective qH.
Nat.Plants, 8, 2022
|
|
7DJK
 
 | | Structure of four truncated and mutated forms of quenching protein | | 分子名称: | CHLORIDE ION, Protein SUPPRESSOR OF QUENCHING 1, chloroplastic, ... | | 著者 | Yu, G.M, Pan, X.W, Li, M. | | 登録日 | 2020-11-20 | | 公開日 | 2022-06-08 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.80145121 Å) | | 主引用文献 | Structure of Arabidopsis SOQ1 lumenal region unveils C-terminal domain essential for negative regulation of photoprotective qH.
Nat.Plants, 8, 2022
|
|
7DJL
 
 | | Structure of four truncated and mutated forms of quenching protein | | 分子名称: | CHLORIDE ION, Protein SUPPRESSOR OF QUENCHING 1, chloroplastic, ... | | 著者 | Yu, G.M, Pan, X.W, Li, M. | | 登録日 | 2020-11-20 | | 公開日 | 2022-06-08 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.96077824 Å) | | 主引用文献 | Structure of Arabidopsis SOQ1 lumenal region unveils C-terminal domain essential for negative regulation of photoprotective qH.
Nat.Plants, 8, 2022
|
|
5NHS
 
 | | The crystal structure of Xanthomonas albilineans N5, N10-methylenetetrahydrofolate dehydrogenase-cyclohydrolase (FolD) | | 分子名称: | 1,2-ETHANEDIOL, Bifunctional protein FolD | | 著者 | Bueno, R.V, Guido, R.V.C, Maluf, F.V, Lima, G.M.A, Oliveira, A.A. | | 登録日 | 2017-03-22 | | 公開日 | 2018-05-16 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (2.11 Å) | | 主引用文献 | The crystal structure of Xanthomonas albilineans N5, N10-methylenetetrahydrofolate dehydrogenase-cyclohydrolase (FolD)
To Be Published
|
|
2LQH
 
 | | NMR structure of FOXO3a transactivation domains (CR2C-CR3) in complex with CBP KIX domain (2b3l conformation) | | 分子名称: | CREB-binding protein, Forkhead box O3 | | 著者 | Wang, F, Marshall, C.B, Yamamoto, K, Li, G.B, Gasmi-Seabrook, G.M.C, Okada, H, Mak, T.W, Ikura, M. | | 登録日 | 2012-03-06 | | 公開日 | 2012-05-16 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structures of KIX domain of CBP in complex with two FOXO3a transactivation domains reveal promiscuity and plasticity in coactivator recruitment.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
5A3C
 
 | | Crystal structure of the ADP-ribosylating sirtuin (SirTM) from Streptococcus pyogenes in complex with NAD | | 分子名称: | 1,2-ETHANEDIOL, GLYCINE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | 著者 | Rack, J.G.M, Morra, R, Barkauskaite, E, Kraehenbuehl, R, Ariza, A, Qu, Y, Ortmayer, M, Leidecker, O, Cameron, D.R, Matic, I, Peleg, A.Y, Leys, D, Traven, A, Ahel, I. | | 登録日 | 2015-05-28 | | 公開日 | 2015-07-29 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2.03 Å) | | 主引用文献 | Identification of a Class of Protein Adp-Ribosylating Sirtuins in Microbial Pathogens.
Mol.Cell, 59, 2015
|
|
5LDT
 
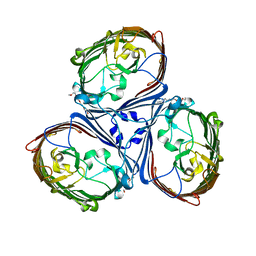 | | Crystal Structures of MOMP from Campylobacter jejuni | | 分子名称: | (HYDROXYETHYLOXY)TRI(ETHYLOXY)OCTANE, CALCIUM ION, MOMP porin | | 著者 | Ferrara, L.G.M, Wallat, G.D, Moynie, L, Naismith, J.H. | | 登録日 | 2016-06-27 | | 公開日 | 2016-10-26 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2.88 Å) | | 主引用文献 | MOMP from Campylobacter jejuni Is a Trimer of 18-Stranded beta-Barrel Monomers with a Ca(2+) Ion Bound at the Constriction Zone.
J.Mol.Biol., 428, 2016
|
|
