2ZZ7
 
 | |
3W4S
 
 | |
3WHB
 
 | |
3WHC
 
 | |
3WQP
 
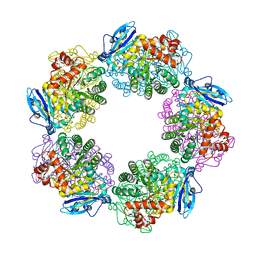 | | Crystal structure of Rubisco T289D mutant from Thermococcus kodakarensis | | 分子名称: | 1,2-ETHANEDIOL, 2-CARBOXYARABINITOL-1,5-DIPHOSPHATE, MAGNESIUM ION, ... | | 著者 | Fujihashi, M, Nishitani, Y, Kiriyama, T, Miki, K. | | 登録日 | 2014-01-29 | | 公開日 | 2015-02-04 | | 最終更新日 | 2023-12-06 | | 実験手法 | X-RAY DIFFRACTION (2.25 Å) | | 主引用文献 | Mutation design of thermophilic Rubisco based on the three-dimensional structure enhances its activity at ambient temperature
to be published
|
|
7E4L
 
 | |
3KDO
 
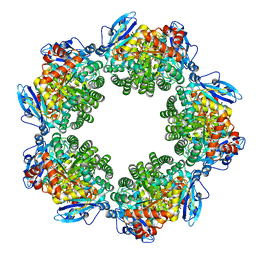 | | Crystal structure of Type III Rubisco SP6 mutant complexed with 2-CABP | | 分子名称: | 2-CARBOXYARABINITOL-1,5-DIPHOSPHATE, MAGNESIUM ION, Ribulose bisphosphate carboxylase | | 著者 | Nishitani, Y, Fujihashi, M, Doi, T, Yoshida, S, Atomi, H, Imanaka, T, Miki, K. | | 登録日 | 2009-10-23 | | 公開日 | 2010-10-06 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.36 Å) | | 主引用文献 | Structure-based catalytic optimization of a type III Rubisco from a hyperthermophile
J.Biol.Chem., 285, 2010
|
|
3KDN
 
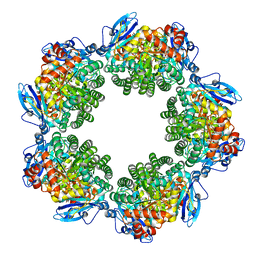 | | Crystal structure of Type III Rubisco SP4 mutant complexed with 2-CABP | | 分子名称: | 2-CARBOXYARABINITOL-1,5-DIPHOSPHATE, MAGNESIUM ION, Ribulose bisphosphate carboxylase | | 著者 | Nishitani, Y, Fujihashi, M, Doi, T, Yoshida, S, Atomi, H, Imanaka, T, Miki, K. | | 登録日 | 2009-10-23 | | 公開日 | 2010-10-06 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.09 Å) | | 主引用文献 | Structure-based catalytic optimization of a type III Rubisco from a hyperthermophile
J.Biol.Chem., 285, 2010
|
|
4XF6
 
 | | myo-inositol 3-kinase bound with its products (ADP and 1D-myo-inositol 3-phosphate) | | 分子名称: | 1,2,3,4,5,6-HEXAHYDROXY-CYCLOHEXANE, 1,2-ETHANEDIOL, ADENOSINE-5'-DIPHOSPHATE, ... | | 著者 | Nagata, R, Fujihashi, M, Miki, K. | | 登録日 | 2014-12-26 | | 公開日 | 2015-06-03 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.08 Å) | | 主引用文献 | Crystal Structure and Product Analysis of an Archaeal myo-Inositol Kinase Reveal Substrate Recognition Mode and 3-OH Phosphorylation
Biochemistry, 54, 2015
|
|
4XF7
 
 | | myo-inositol 3-kinase bound with its substrates (AMPPCP and myo-inositol) | | 分子名称: | 1,2,3,4,5,6-HEXAHYDROXY-CYCLOHEXANE, Carbohydrate/pyrimidine kinase, PfkB family, ... | | 著者 | Nagata, R, Fujihashi, M, Miki, K. | | 登録日 | 2014-12-26 | | 公開日 | 2015-06-03 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.93 Å) | | 主引用文献 | Crystal Structure and Product Analysis of an Archaeal myo-Inositol Kinase Reveal Substrate Recognition Mode and 3-OH Phosphorylation
Biochemistry, 54, 2015
|
|
1WMF
 
 | | Crystal Structure of alkaline serine protease KP-43 from Bacillus sp. KSM-KP43 (oxidized form, 1.73 angstrom) | | 分子名称: | 1,4-DIETHYLENE DIOXIDE, CALCIUM ION, GLYCEROL, ... | | 著者 | Nonaka, T, Fujihashi, M, Kita, A, Saeki, K, Ito, S, Horikoshi, K, Miki, K. | | 登録日 | 2004-07-08 | | 公開日 | 2004-09-14 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (1.73 Å) | | 主引用文献 | The Crystal Structure of an Oxidatively Stable Subtilisin-like Alkaline Serine Protease, KP-43, with a C-terminal {beta}-Barrel Domain
J.Biol.Chem., 279, 2004
|
|
1WME
 
 | | Crystal Structure of alkaline serine protease KP-43 from Bacillus sp. KSM-KP43 (1.50 angstrom, 293 K) | | 分子名称: | CALCIUM ION, protease | | 著者 | Nonaka, T, Fujihashi, M, Kita, A, Saeki, K, Ito, S, Horikoshi, K, Miki, K. | | 登録日 | 2004-07-08 | | 公開日 | 2004-09-14 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | The Crystal Structure of an Oxidatively Stable Subtilisin-like Alkaline Serine Protease, KP-43, with a C-terminal {beta}-Barrel Domain
J.Biol.Chem., 279, 2004
|
|
1WMD
 
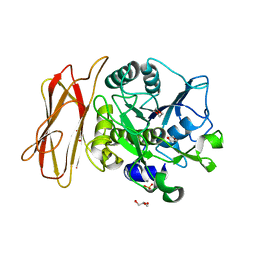 | | Crystal Structure of alkaline serine protease KP-43 from Bacillus sp. KSM-KP43 (1.30 angstrom, 100 K) | | 分子名称: | 1,4-DIETHYLENE DIOXIDE, CALCIUM ION, GLYCEROL, ... | | 著者 | Nonaka, T, Fujihashi, M, Kita, A, Saeki, K, Ito, S, Horikoshi, K, Miki, K. | | 登録日 | 2004-07-08 | | 公開日 | 2004-09-14 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (1.3 Å) | | 主引用文献 | The Crystal Structure of an Oxidatively Stable Subtilisin-like Alkaline Serine Protease, KP-43, with a C-terminal {beta}-Barrel Domain
J.Biol.Chem., 279, 2004
|
|
1UD6
 
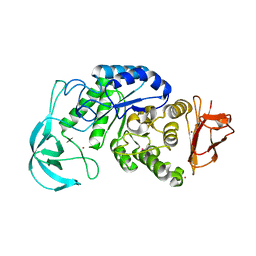 | | Crystal structure of AmyK38 with potassium ion | | 分子名称: | POTASSIUM ION, amylase | | 著者 | Nonaka, T, Fujihashi, M, Kita, A, Hagihara, H, Ozaki, K, Ito, S, Miki, K. | | 登録日 | 2003-04-28 | | 公開日 | 2003-07-22 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Crystal structure of calcium-free alpha-amylase from Bacillus sp. strain KSM-K38 (AmyK38) and its sodium ion binding sites
J.Biol.Chem., 278, 2003
|
|
1UD3
 
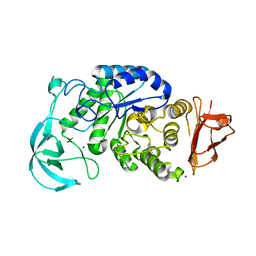 | | Crystal structure of AmyK38 N289H mutant | | 分子名称: | SODIUM ION, amylase | | 著者 | Nonaka, T, Fujihashi, M, Kita, A, Hagihara, H, Ozaki, K, Ito, S, Miki, K. | | 登録日 | 2003-04-28 | | 公開日 | 2003-07-22 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (2.15 Å) | | 主引用文献 | Crystal structure of calcium-free alpha-amylase from Bacillus sp. strain KSM-K38 (AmyK38) and its sodium ion binding sites
J.Biol.Chem., 278, 2003
|
|
1UD8
 
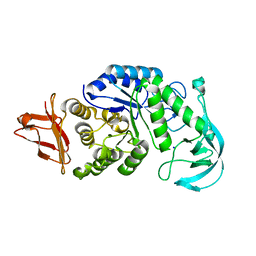 | | Crystal structure of AmyK38 with lithium ion | | 分子名称: | SODIUM ION, amylase | | 著者 | Nonaka, T, Fujihashi, M, Kita, A, Hagihara, H, Ozaki, K, Ito, S, Miki, K. | | 登録日 | 2003-04-28 | | 公開日 | 2003-07-22 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (2.88 Å) | | 主引用文献 | Crystal structure of calcium-free alpha-amylase from Bacillus sp. strain KSM-K38 (AmyK38) and its sodium ion binding sites
J.Biol.Chem., 278, 2003
|
|
1UD5
 
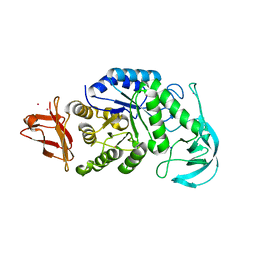 | | Crystal structure of AmyK38 with rubidium ion | | 分子名称: | RUBIDIUM ION, SODIUM ION, amylase | | 著者 | Nonaka, T, Fujihashi, M, Kita, A, Hagihara, H, Ozaki, K, Ito, S, Miki, K. | | 登録日 | 2003-04-28 | | 公開日 | 2003-07-22 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Crystal structure of calcium-free alpha-amylase from Bacillus sp. strain KSM-K38 (AmyK38) and its sodium ion binding sites
J.Biol.Chem., 278, 2003
|
|
1UD4
 
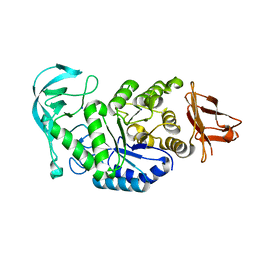 | | Crystal structure of calcium free alpha amylase from Bacillus sp. strain KSM-K38 (AmyK38, in calcium containing solution) | | 分子名称: | SODIUM ION, amylase | | 著者 | Nonaka, T, Fujihashi, M, Kita, A, Hagihara, H, Ozaki, K, Ito, S, Miki, K. | | 登録日 | 2003-04-28 | | 公開日 | 2003-07-22 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (2.15 Å) | | 主引用文献 | Crystal structure of calcium-free alpha-amylase from Bacillus sp. strain KSM-K38 (AmyK38) and its sodium ion binding sites
J.Biol.Chem., 278, 2003
|
|
1UD2
 
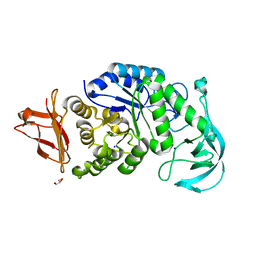 | | Crystal structure of calcium-free alpha-amylase from Bacillus sp. strain KSM-K38 (AmyK38) | | 分子名称: | GLYCEROL, SODIUM ION, amylase | | 著者 | Nonaka, T, Fujihashi, M, Kita, A, Hagihara, H, Ozaki, K, Ito, S, Miki, K. | | 登録日 | 2003-04-28 | | 公開日 | 2003-07-22 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.13 Å) | | 主引用文献 | Crystal structure of calcium-free alpha-amylase from Bacillus sp. strain KSM-K38 (AmyK38) and its sodium ion binding sites
J.Biol.Chem., 278, 2003
|
|
5X0J
 
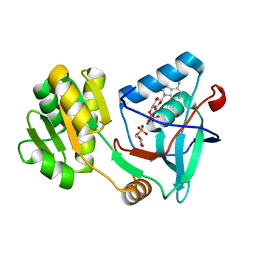 | | Free serine kinase (E30Q mutant) in complex with phosphoserine and AMP | | 分子名称: | ADENOSINE MONOPHOSPHATE, Free serine kinase, MAGNESIUM ION, ... | | 著者 | Nagata, R, Fujihashi, M, Miki, K. | | 登録日 | 2017-01-20 | | 公開日 | 2017-04-12 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.43 Å) | | 主引用文献 | Structural Study on the Reaction Mechanism of a Free Serine Kinase Involved in Cysteine Biosynthesis
ACS Chem. Biol., 12, 2017
|
|
5X0F
 
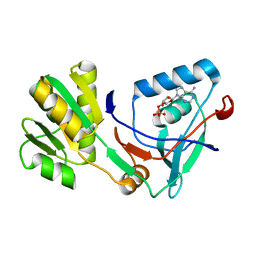 | |
5X0K
 
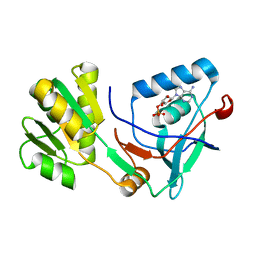 | |
5YSQ
 
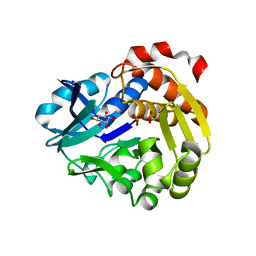 | | Sulfate-complex structure of a pyrophosphate-dependent kinase in the ribokinase family provides insight into the donor-binding mode | | 分子名称: | 1,2,3,4,5,6-HEXAHYDROXY-CYCLOHEXANE, SULFATE ION, TM0415 | | 著者 | Nagata, R, Fujihashi, M, Miki, K. | | 登録日 | 2017-11-14 | | 公開日 | 2018-05-16 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.47 Å) | | 主引用文献 | Identification of a pyrophosphate-dependent kinase and its donor selectivity determinants.
Nat Commun, 9, 2018
|
|
5YSP
 
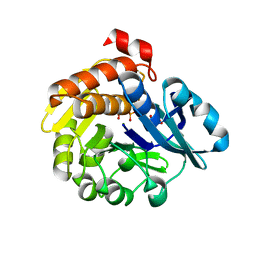 | | Pyrophosphate-dependent kinase in the ribokinase family complexed with a pyrophosphate analog and myo-inositol | | 分子名称: | 1,2,3,4,5,6-HEXAHYDROXY-CYCLOHEXANE, MAGNESIUM ION, METHYLENEDIPHOSPHONIC ACID, ... | | 著者 | Nagata, R, Fujihashi, M, Miki, K. | | 登録日 | 2017-11-14 | | 公開日 | 2018-05-16 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Identification of a pyrophosphate-dependent kinase and its donor selectivity determinants.
Nat Commun, 9, 2018
|
|
5X0G
 
 | | Free serine kinase (E30A mutant) in complex with ADP | | 分子名称: | 2-hydroxy-3-[4-(2-hydroxy-3-sulfopropyl)piperazin-1-yl]propane-1-sulfonic acid, ADENOSINE MONOPHOSPHATE, ADENOSINE-5'-DIPHOSPHATE, ... | | 著者 | Nagata, R, Fujihashi, M, Miki, K. | | 登録日 | 2017-01-20 | | 公開日 | 2017-04-12 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Structural Study on the Reaction Mechanism of a Free Serine Kinase Involved in Cysteine Biosynthesis
ACS Chem. Biol., 12, 2017
|
|
