8U0I
 
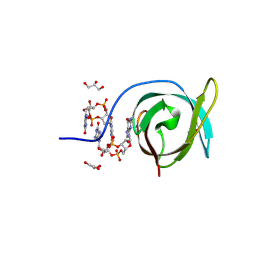 | | Crystal structure of PA0012 complexed with cyclic-di-GMP from Pseudomonas aeruginosa | | 分子名称: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), GLYCEROL, PilZ domain-containing protein | | 著者 | Hammons, N.A, Schnicker, N.J, Fuentes, E.J. | | 登録日 | 2023-08-29 | | 公開日 | 2024-09-04 | | 実験手法 | X-RAY DIFFRACTION (1.541 Å) | | 主引用文献 | Crystal structure of PA0012 complexed with cyclic-di-GMP from Pseudomonas aeruginosa
To Be Published
|
|
7JL6
 
 | |
1LQ7
 
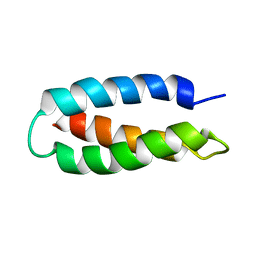 | | De Novo Designed Protein Model of Radical Enzymes | | 分子名称: | Alpha3W | | 著者 | Dai, Q.-H, Tommos, C, Fuentes, E.J, Blomberg, M, Dutton, P.L, Wand, A.J. | | 登録日 | 2002-05-09 | | 公開日 | 2002-06-05 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structure of a De Novo Designed Protein Model of Radical Enzymes
J.Am.Chem.Soc., 124, 2002
|
|
3KZD
 
 | |
3KZE
 
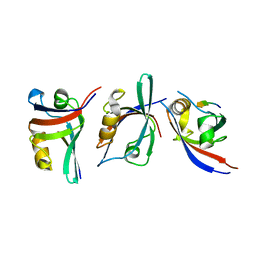 | |
4GVD
 
 | | Crystal Structure of T-cell Lymphoma Invasion and Metastasis-1 PDZ in complex with Syndecan1 Peptide | | 分子名称: | 5-(DIMETHYLAMINO)-1-NAPHTHALENESULFONIC ACID(DANSYL ACID), CHLORIDE ION, SODIUM ION, ... | | 著者 | Liu, X, Shepherd, T.R, Murray, A.M, Xu, Z, Fuentes, E.J. | | 登録日 | 2012-08-30 | | 公開日 | 2013-03-13 | | 最終更新日 | 2024-10-16 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | The structure of the Tiam1 PDZ domain/ phospho-syndecan1 complex reveals a ligand conformation that modulates protein dynamics.
Structure, 21, 2013
|
|
4GVC
 
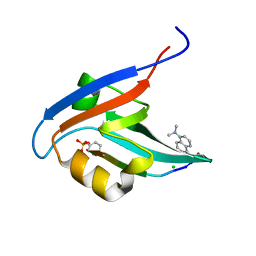 | | Crystal Structure of T-cell Lymphoma Invasion and Metastasis-1 PDZ in complex with phosphorylated Syndecan1 Peptide | | 分子名称: | 5-(DIMETHYLAMINO)-1-NAPHTHALENESULFONIC ACID(DANSYL ACID), CHLORIDE ION, SODIUM ION, ... | | 著者 | Liu, X, Shepherd, T.R, Murray, A.M, Xu, Z, Fuentes, E.J. | | 登録日 | 2012-08-30 | | 公開日 | 2013-03-13 | | 最終更新日 | 2023-12-06 | | 実験手法 | X-RAY DIFFRACTION (1.54 Å) | | 主引用文献 | The structure of the Tiam1 PDZ domain/ phospho-syndecan1 complex reveals a ligand conformation that modulates protein dynamics.
Structure, 21, 2013
|
|
6MYE
 
 | | Crystal structure of human Scribble PDZ1 domain in complex with internal PDZ binding motif of Src homology 3 domain-containing guanine nucleotide exchange factor (SGEF) | | 分子名称: | FORMIC ACID, Protein scribble homolog, Rho guanine nucleotide exchange factor 26, ... | | 著者 | Sun, Y.J, Hou, T, Gakhar, L, Fuentes, E.J. | | 登録日 | 2018-11-01 | | 公開日 | 2019-04-10 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.1 Å) | | 主引用文献 | SGEF forms a complex with Scribble and Dlg1 and regulates epithelial junctions and contractility.
J.Cell Biol., 218, 2019
|
|
6MYF
 
 | |
6NID
 
 | |
6NEK
 
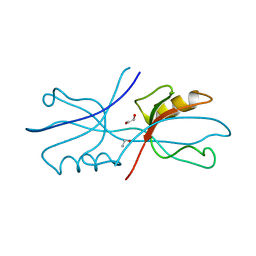 | |
6NH9
 
 | |
4NXP
 
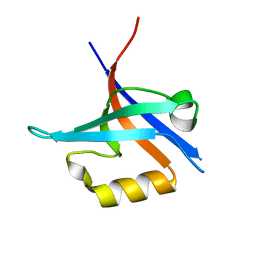 | | Crystal Structure of Free T-cell Lymphoma Invasion and Metastasis-1 PDZ Domain Quadruple Mutant (QM) | | 分子名称: | T-lymphoma invasion and metastasis-inducing protein 1 | | 著者 | Liu, X, Speckhard, D.C, Shepherd, T.R, Hengel, S.R, Fuentes, E.J. | | 登録日 | 2013-12-09 | | 公開日 | 2015-05-13 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Distinct Roles for Conformational Dynamics in Protein-Ligand Interactions.
Structure, 24, 2016
|
|
4NXQ
 
 | | Crystal Structure of T-cell Lymphoma Invasion and Metastasis-1 PDZ Domain Quadruple Mutant (QM) in Complex With Caspr4 Peptide | | 分子名称: | Contactin-associated protein-like 4 peptide, T-lymphoma invasion and metastasis-inducing protein 1 | | 著者 | Liu, X, Speckhard, D.C, Shepherd, T.R, Hengel, S.R, Fuentes, E.J. | | 登録日 | 2013-12-09 | | 公開日 | 2015-05-13 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Distinct Roles for Conformational Dynamics in Protein-Ligand Interactions.
Structure, 24, 2016
|
|
4K2O
 
 | |
4K2P
 
 | |
4NXR
 
 | | Crystal Structure of T-cell Lymphoma Invasion and Metastasis-1 PDZ Domain Quadruple Mutant (QM) in Complex With Neurexin-1 Peptide | | 分子名称: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, 5-(DIMETHYLAMINO)-1-NAPHTHALENESULFONIC ACID(DANSYL ACID), Neurexin-2-beta Peptide, ... | | 著者 | Liu, X, Speckhard, D.C, Shepherd, T.R, Hengel, S.R, Fuentes, E.J. | | 登録日 | 2013-12-09 | | 公開日 | 2015-05-13 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Distinct Roles for Conformational Dynamics in Protein-Ligand Interactions.
Structure, 24, 2016
|
|
