3NYE
 
 | |
6A4I
 
 | | Crystal Structure of human TDO inhibitor complex | | Descriptor: | 1-(6-chloro-1H-indazol-4-yl)cyclohexan-1-ol, CITRIC ACID, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Fu, G, Wang, J, Luo, G, Wu, G, Qian, K. | | Deposit date: | 2018-06-20 | | Release date: | 2018-07-18 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Crystal Structure of human TDO inhibitor complex
To Be Published
|
|
3SM8
 
 | |
2ZJT
 
 | |
3NYC
 
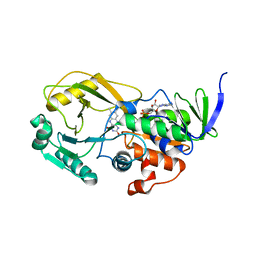 | | Crystal Structure of Pseudomonas aeruginosa D-Arginine Dehydrogenase | | Descriptor: | (2E)-5-[(diaminomethylidene)amino]-2-iminopentanoic acid, D-Arginine Dehydrogenase, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Fu, G, Weber, I.T. | | Deposit date: | 2010-07-14 | | Release date: | 2010-09-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.06 Å) | | Cite: | Conformational changes and substrate recognition in Pseudomonas aeruginosa D-arginine dehydrogenase.
Biochemistry, 49, 2010
|
|
3NYF
 
 | |
3EDQ
 
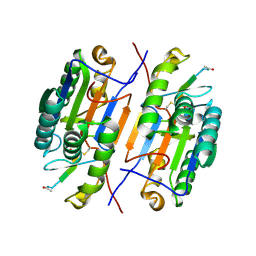 | | Crystal structure of Caspase-3 with inhibitor AC-LDESD-CHO | | Descriptor: | AC-LDESD-CHO peptide, Caspase-3 | | Authors: | Fu, G. | | Deposit date: | 2008-09-03 | | Release date: | 2008-10-28 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Structural basis for executioner caspase recognition of P5 position in substrates.
Apoptosis, 13, 2008
|
|
2VHA
 
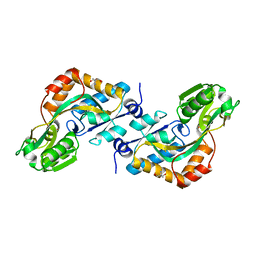 | | DEBP | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, GLUTAMIC ACID, PERIPLASMIC BINDING TRANSPORT PROTEIN | | Authors: | Hu, Y.L, Fan, C.-P, Fu, G.S, Zhu, D.Y, Jin, Q, Wang, D.-C. | | Deposit date: | 2007-11-20 | | Release date: | 2008-07-08 | | Last modified: | 2019-05-22 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Crystal Structure of a Glutamate/Aspartate Binding Protein Complexed with a Glutamate Molecule: Structural Basis of Ligand Specificity at Atomic Resolution.
J.Mol.Biol., 382, 2008
|
|
3GJS
 
 | | Caspase-3 Binds Diverse P4 Residues in Peptides | | Descriptor: | Ac-YVAD-Cho inhibitor, Caspase-3 subunit p12, Caspase-3 subunit p17 | | Authors: | Fang, B, Fu, G, Agniswamy, J, Harrison, R.W, Weber, I.T. | | Deposit date: | 2009-03-09 | | Release date: | 2009-03-24 | | Last modified: | 2011-08-17 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Caspase-3 binds diverse P4 residues in peptides as revealed by crystallography and structural modeling.
Apoptosis, 14, 2009
|
|
3GJR
 
 | | Caspase-3 Binds Diverse P4 Residues in Peptides | | Descriptor: | Caspase-3 subunit p12, Caspase-3 subunit p17, GLYCEROL, ... | | Authors: | Fang, B, Fu, G, Agniswamy, J, Harrison, R.W, Weber, I.T. | | Deposit date: | 2009-03-09 | | Release date: | 2009-03-24 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Caspase-3 binds diverse P4 residues in peptides as revealed by crystallography and structural modeling.
Apoptosis, 14, 2009
|
|
3GJT
 
 | | Caspase-3 Binds Diverse P4 Residues in Peptides | | Descriptor: | Caspase-3 subunit p12, Caspase-3 subunit p17, peptide inhibitor | | Authors: | Fang, B, Fu, G, Agniswamy, J, Harrison, R.W, Weber, I.T. | | Deposit date: | 2009-03-09 | | Release date: | 2009-03-24 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Caspase-3 binds diverse P4 residues in peptides as revealed by crystallography and structural modeling.
Apoptosis, 14, 2009
|
|
3GJQ
 
 | | Caspase-3 Binds Diverse P4 Residues in Peptides | | Descriptor: | Caspase-3 subunit p12, Caspase-3 subunit p17, peptide inhibitor | | Authors: | Fang, B, Fu, G, Agniswamy, J, Harrison, R.W, Weber, I.T. | | Deposit date: | 2009-03-09 | | Release date: | 2009-03-24 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Caspase-3 binds diverse P4 residues in peptides as revealed by crystallography and structural modeling.
Apoptosis, 14, 2009
|
|
5U36
 
 | | Crystal Structure Of A Mutant M. Jannashii Tyrosyl-tRNA Synthetase | | Descriptor: | Tyrosine--tRNA ligase | | Authors: | Luo, X, Fu, G, Zhu, X, Wilson, I.A, Wang, F. | | Deposit date: | 2016-12-01 | | Release date: | 2017-06-07 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.03 Å) | | Cite: | Genetically encoding phosphotyrosine and its nonhydrolyzable analog in bacteria.
Nat. Chem. Biol., 13, 2017
|
|
