6OB5
 
 | | Computationally-designed, modular sense/response system (S3-2D) | | Descriptor: | Ankyrin Repeat Domain (AR), S3-2D variant, FARNESYL DIPHOSPHATE, ... | | Authors: | Thompson, M.C, Glasgow, A.A, Huang, Y.M, Fraser, J.S, Kortemme, T. | | Deposit date: | 2019-03-19 | | Release date: | 2019-12-04 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.208 Å) | | Cite: | Computational design of a modular protein sense-response system.
Science, 366, 2019
|
|
8SOV
 
 | | Proteinase K Multiconformer Model at 353K | | Descriptor: | ALA-ALA-ALA-SER-VAL-LYS, CALCIUM ION, Proteinase K, ... | | Authors: | Du, S, Wankowicz, S, Yabukarski, F, Doukov, T, Herschlag, D, Fraser, J.S. | | Deposit date: | 2023-04-30 | | Release date: | 2023-08-09 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.291 Å) | | Cite: | Refinement of multiconformer ensemble models from multi-temperature X-ray diffraction data.
Methods Enzymol., 688, 2023
|
|
8SPL
 
 | | Proteinase K Multiconformer Model at 343K | | Descriptor: | ALA-ALA-ALA-SER-VAL-LYS, CALCIUM ION, Proteinase K, ... | | Authors: | Du, S, Wankowicz, S, Yabukarski, F, Doukov, T, Herschlag, D, Fraser, J.S. | | Deposit date: | 2023-05-03 | | Release date: | 2023-08-09 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.21 Å) | | Cite: | Refinement of multiconformer ensemble models from multi-temperature X-ray diffraction data.
Methods Enzymol., 688, 2023
|
|
8SOG
 
 | | Proteinase K Multiconformer Model at 313K | | Descriptor: | CALCIUM ION, Proteinase K, SULFATE ION | | Authors: | Du, S, Wankowicz, S, Yabukarski, F, Doukov, T, Herschlag, D, Fraser, J.S. | | Deposit date: | 2023-04-28 | | Release date: | 2023-08-09 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.13 Å) | | Cite: | Refinement of multiconformer ensemble models from multi-temperature X-ray diffraction data.
Methods Enzymol., 688, 2023
|
|
8SOU
 
 | | Proteinase K Multiconformer Model at 363K | | Descriptor: | ALA-ALA-ALA-SER-VAL-LYS, CALCIUM ION, Proteinase K, ... | | Authors: | Du, S, Wankowicz, S, Yabukarski, F, Doukov, T, Herschlag, D, Fraser, J.S. | | Deposit date: | 2023-04-30 | | Release date: | 2023-08-09 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Refinement of multiconformer ensemble models from multi-temperature X-ray diffraction data.
Methods Enzymol., 688, 2023
|
|
8SQV
 
 | | Proteinase K Multiconformer Model at 333K | | Descriptor: | CALCIUM ION, Proteinase K, SULFATE ION | | Authors: | Du, S, Wankowicz, S, Yabukarski, F, Doukov, T, Herschlag, D, Fraser, J.S. | | Deposit date: | 2023-05-04 | | Release date: | 2023-08-09 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.22 Å) | | Cite: | Refinement of multiconformer ensemble models from multi-temperature X-ray diffraction data.
Methods Enzymol., 688, 2023
|
|
8GCA
 
 | | Mouse acidic mammalian chitinase, catalytic domain in complex with N,N',N''-triacetylchitotriose at pH 4.74 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Acidic mammalian chitinase, ... | | Authors: | Diaz, R.E, Fraser, J.S. | | Deposit date: | 2023-03-01 | | Release date: | 2023-03-15 | | Last modified: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural characterization of ligand binding and pH-specific enzymatic activity of mouse Acidic Mammalian Chitinase.
Biorxiv, 2024
|
|
6X3C
 
 | | Crystal structure of streptogramin A acetyltransferase VatA from Staphylococcus aureus in complex with streptogramin analog F1037 (47) | | Descriptor: | (3R,4R,5E,10E,12E,14S,16R,26aR)-16-fluoro-14-hydroxy-12-methyl-3-(propan-2-yl)-4-(prop-2-en-1-yl)-3,4,8,9,14,15,16,17,24,25,26,26a-dodecahydro-1H,7H,22H-21,18-(azeno)pyrrolo[2,1-c][1,8,4,19]dioxadiazacyclotetracosine-1,7,22-trione, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Chaires, H.A, Fraser, J.S. | | Deposit date: | 2020-05-21 | | Release date: | 2020-11-18 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Synthetic group A streptogramin antibiotics that overcome Vat resistance.
Nature, 586, 2020
|
|
3J9J
 
 | | Structure of the capsaicin receptor, TRPV1, determined by single particle electron cryo-microscopy | | Descriptor: | Transient receptor potential cation channel subfamily V member 1 | | Authors: | Wang, R.Y.-R, Barad, B.A, Fraser, J.S, DiMaio, F. | | Deposit date: | 2015-02-02 | | Release date: | 2015-09-02 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (3.275 Å) | | Cite: | EMRinger: side chain-directed model and map validation for 3D cryo-electron microscopy.
Nat.Methods, 12, 2015
|
|
8FOR
 
 | |
8FMC
 
 | |
8FMD
 
 | |
8FOQ
 
 | |
8FME
 
 | |
8FOS
 
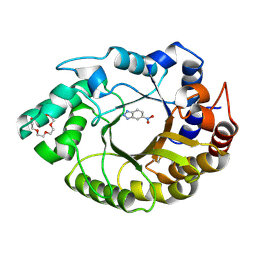 | |
7S1G
 
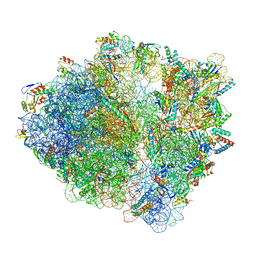 | | wild-type Escherichia coli stalled ribosome with antibiotic linezolid | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Young, I.D, Stojkovic, V, Tsai, K, Lee, D.J, Fraser, J.S, Galonic Fujimori, D. | | Deposit date: | 2021-09-02 | | Release date: | 2021-11-17 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (2.48 Å) | | Cite: | Structural basis for context-specific inhibition of translation by oxazolidinone antibiotics.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7S1H
 
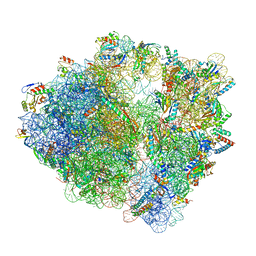 | | Wild-type Escherichia coli ribosome with antibiotic linezolid | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Young, I.D, Stojkovic, V, Tsai, K, Lee, D.J, Fraser, J.S, Galonic Fujimori, D. | | Deposit date: | 2021-09-02 | | Release date: | 2021-11-17 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (2.35 Å) | | Cite: | Structural basis for context-specific inhibition of translation by oxazolidinone antibiotics.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7KQP
 
 | | Crystal structure of SARS-CoV-2 NSP3 macrodomain in complex with ADP-ribose (P43 crystal form) | | Descriptor: | Non-structural protein 3, [(2R,3S,4R,5R)-5-(6-AMINOPURIN-9-YL)-3,4-DIHYDROXY-OXOLAN-2-YL]METHYL [HYDROXY-[[(2R,3S,4R,5S)-3,4,5-TRIHYDROXYOXOLAN-2-YL]METHOXY]PHOSPHORYL] HYDROGEN PHOSPHATE | | Authors: | Correy, G.J, Young, I.D, Thompson, M.C, Fraser, J.S. | | Deposit date: | 2020-11-17 | | Release date: | 2020-12-09 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (0.88 Å) | | Cite: | Fragment binding to the Nsp3 macrodomain of SARS-CoV-2 identified through crystallographic screening and computational docking.
Sci Adv, 7, 2021
|
|
7KQW
 
 | | Crystal structure of SARS-CoV-2 NSP3 macrodomain (C2 crystal form, methylated) | | Descriptor: | Non-structural protein 3 | | Authors: | Correy, G.J, Young, I.D, Thompson, M.C, Fraser, J.S. | | Deposit date: | 2020-11-17 | | Release date: | 2020-12-09 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (0.93 Å) | | Cite: | Fragment binding to the Nsp3 macrodomain of SARS-CoV-2 identified through crystallographic screening and computational docking.
Sci Adv, 7, 2021
|
|
7KR1
 
 | | Crystal structure of SARS-CoV-2 NSP3 macrodomain (C2 crystal form, 310 K) | | Descriptor: | Non-structural protein 3 | | Authors: | Correy, G.J, Young, I.D, Thompson, M.C, Fraser, J.S. | | Deposit date: | 2020-11-18 | | Release date: | 2020-12-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Fragment binding to the Nsp3 macrodomain of SARS-CoV-2 identified through crystallographic screening and computational docking.
Sci Adv, 7, 2021
|
|
7KR0
 
 | | Crystal structure of SARS-CoV-2 NSP3 macrodomain (C2 crystal form, 100 K) | | Descriptor: | Non-structural protein 3 | | Authors: | Correy, G.J, Young, I.D, Thompson, M.C, Fraser, J.S. | | Deposit date: | 2020-11-18 | | Release date: | 2020-12-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (0.77 Å) | | Cite: | Fragment binding to the Nsp3 macrodomain of SARS-CoV-2 identified through crystallographic screening and computational docking.
Sci Adv, 7, 2021
|
|
6O2S
 
 | | Deacetylated Microtubules | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Eshun-Wilson, L, Zhang, R, Portran, D, Nachury, M.V, Toso, D, Lohr, T, Vendruscolo, M, Bonomi, M, Fraser, J.S, Nogales, E. | | Deposit date: | 2019-02-24 | | Release date: | 2019-05-22 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Effects of alpha-tubulin acetylation on microtubule structure and stability.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6O2T
 
 | | Acetylated Microtubules | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Eshun-Wilson, L, Zhang, R, Portran, D, Nachury, M.V, Toso, D, Lohr, T, Vendruscolo, M, Bonomi, M, Fraser, J.S, Nogales, E. | | Deposit date: | 2019-02-24 | | Release date: | 2019-05-22 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Effects of alpha-tubulin acetylation on microtubule structure and stability.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6O2R
 
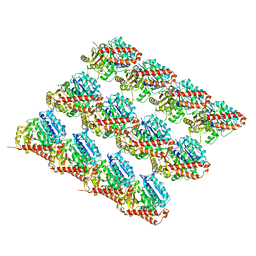 | | Deacetylated Microtubules | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Eshun-Wilson, L, Zhang, R, Portran, D, Nachury, M.V, Toso, D, Lohr, T, Vendruscolo, M, Bonomi, M, Fraser, J.S, Nogales, E. | | Deposit date: | 2019-02-24 | | Release date: | 2019-05-22 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Effects of alpha-tubulin acetylation on microtubule structure and stability.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6O2Q
 
 | | Acetylated Microtubules | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Eshun-Wilson, L, Zhang, R, Portran, D, Nachury, M.V, Toso, D, Lohr, T, Vendruscolo, M, Bonomi, M, Fraser, J.S, Nogales, E. | | Deposit date: | 2019-02-24 | | Release date: | 2019-05-22 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Effects of alpha-tubulin acetylation on microtubule structure and stability.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
