1ZC8
 
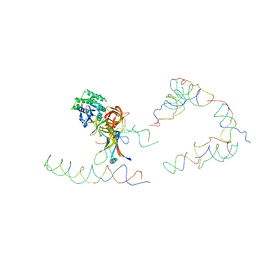 | | Coordinates of tmRNA, SmpB, EF-Tu and h44 fitted into Cryo-EM map of the 70S ribosome and tmRNA complex | | 分子名称: | Elongation factor Tu, H2 16S rRNA, H2b d mRNA, ... | | 著者 | Valle, M, Gillet, R, Kaur, S, Henne, A, Ramakrishnan, V, Frank, J. | | 登録日 | 2005-04-11 | | 公開日 | 2005-04-19 | | 最終更新日 | 2024-02-14 | | 実験手法 | ELECTRON MICROSCOPY (13 Å) | | 主引用文献 | Visualizing tmRNA Entry into a Stalled Ribosome
Science, 300, 2003
|
|
3IZ4
 
 | |
7SYN
 
 | | Structure of the HCV IRES bound to the 40S ribosomal subunit, head opening. Structure 8(delta dII) | | 分子名称: | 18S rRNA, 40S ribosomal protein S2, HCV IRES, ... | | 著者 | Brown, Z.P, Abaeva, I.S, De, S, Hellen, C.U.T, Pestova, T.V, Frank, J. | | 登録日 | 2021-11-25 | | 公開日 | 2022-07-13 | | 実験手法 | ELECTRON MICROSCOPY (4 Å) | | 主引用文献 | Molecular architecture of 40S initiation complexes on the Hepatitis C virus IRES: from ribosomal attachment to eIF5B-mediated reorientation of initiator tRNA
To Be Published
|
|
7SYG
 
 | | Structure of the HCV IRES binding to the 40S ribosomal subunit, closed conformation. Structure 1(delta dII) | | 分子名称: | 18S rRNA, 40S ribosomal protein S2, 40S ribosomal protein S24, ... | | 著者 | Brown, Z.P, Abaeva, I.S, De, S, Hellen, C.U.T, Pestova, T.V, Frank, J. | | 登録日 | 2021-11-25 | | 公開日 | 2022-07-13 | | 最終更新日 | 2024-02-28 | | 実験手法 | ELECTRON MICROSCOPY (4.3 Å) | | 主引用文献 | Molecular architecture of 40S initiation complexes on the Hepatitis C virus IRES: from ribosomal attachment to eIF5B-mediated reorientation of initiator tRNA
To Be Published
|
|
7SYM
 
 | | Structure of the HCV IRES bound to the 40S ribosomal subunit, head opening. Structure 7(delta dII) | | 分子名称: | 18S rRNA, 40S ribosomal protein S2, 40S ribosomal protein S21, ... | | 著者 | Brown, Z.P, Abaeva, I.S, De, S, Hellen, C.U.T, Pestova, T.V, Frank, J. | | 登録日 | 2021-11-25 | | 公開日 | 2022-07-13 | | 最終更新日 | 2024-02-28 | | 実験手法 | ELECTRON MICROSCOPY (4.8 Å) | | 主引用文献 | Comprehensive structural overview of the HCV IRES-mediated translation initiation pathway
To Be Published
|
|
6OUO
 
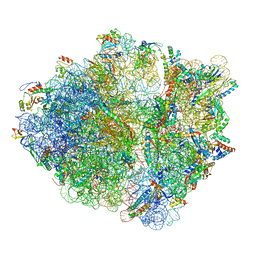 | | RF2 accommodated state bound 70S complex at long incubation time | | 分子名称: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | 著者 | Fu, Z, Indrisiunaite, G, Kaledhonkar, S, Shah, B, Sun, M, Chen, B, Grassucci, R.A, Ehrenberg, M, Frank, J. | | 登録日 | 2019-05-05 | | 公開日 | 2019-06-19 | | 最終更新日 | 2024-03-20 | | 実験手法 | ELECTRON MICROSCOPY (3.7 Å) | | 主引用文献 | The structural basis for release-factor activation during translation termination revealed by time-resolved cryogenic electron microscopy.
Nat Commun, 10, 2019
|
|
7SYH
 
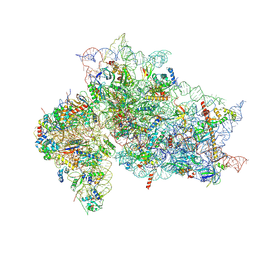 | | Structure of the HCV IRES binding to the 40S ribosomal subunit, closed conformation. Structure 2(delta dII) | | 分子名称: | 18S rRNA, 40S ribosomal protein S21, 40S ribosomal protein S24, ... | | 著者 | Brown, Z.P, Abaeva, I.S, De, S, Hellen, C.U.T, Pestova, T.V, Frank, J. | | 登録日 | 2021-11-25 | | 公開日 | 2022-07-13 | | 実験手法 | ELECTRON MICROSCOPY (4.6 Å) | | 主引用文献 | Molecular architecture of 40S initiation complexes on the Hepatitis C virus IRES: from ribosomal attachment to eIF5B-mediated reorientation of initiator tRNA
To Be Published
|
|
6OSQ
 
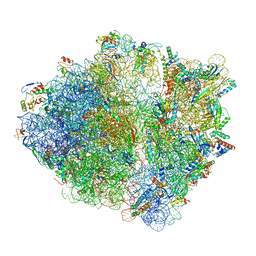 | | RF1 accommodated state bound Release complex 70S at long incubation time point | | 分子名称: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | 著者 | Fu, Z, Indrisiunaite, G, Kaledhonkar, S, Shah, B, Sun, M, Chen, B, Grassucci, R.A, Ehrenberg, M, Frank, J. | | 登録日 | 2019-05-02 | | 公開日 | 2019-06-26 | | 最終更新日 | 2020-01-08 | | 実験手法 | ELECTRON MICROSCOPY (3.5 Å) | | 主引用文献 | The structural basis for release-factor activation during translation termination revealed by time-resolved cryogenic electron microscopy.
Nat Commun, 10, 2019
|
|
5WEN
 
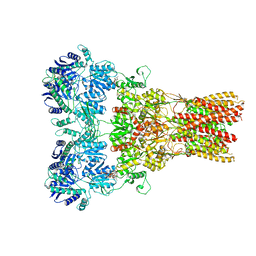 | | GluA2 bound to GSG1L in digitonin, state 2 | | 分子名称: | Digitonin, Glutamate receptor 2,Germ cell-specific gene 1-like protein | | 著者 | Twomey, E.C, Yelshanskaya, M.V, Grassucci, R.A, Frank, J, Sobolevsky, A.I. | | 登録日 | 2017-07-10 | | 公開日 | 2017-08-02 | | 最終更新日 | 2021-08-25 | | 実験手法 | ELECTRON MICROSCOPY (6.8 Å) | | 主引用文献 | Channel opening and gating mechanism in AMPA-subtype glutamate receptors.
Nature, 549, 2017
|
|
3JBO
 
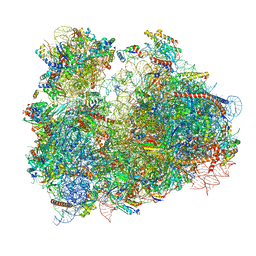 | | Cryo-electron microscopy reconstruction of the Plasmodium falciparum 80S ribosome bound to P/E-tRNA | | 分子名称: | 18S ribosomal RNA, 28S ribosomal RNA, 40S ribosomal protein eS1, ... | | 著者 | Sun, M, Li, W, Blomqvist, K, Das, S, Hashem, Y, Dvorin, J.D, Frank, J. | | 登録日 | 2015-09-16 | | 公開日 | 2015-10-14 | | 最終更新日 | 2024-02-21 | | 実験手法 | ELECTRON MICROSCOPY (5.8 Å) | | 主引用文献 | Dynamical features of the Plasmodium falciparum ribosome during translation.
Nucleic Acids Res., 43, 2015
|
|
3JBP
 
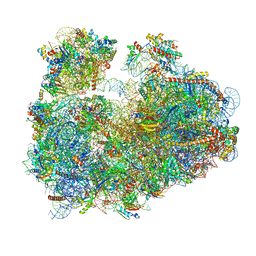 | | Cryo-electron microscopy reconstruction of the Plasmodium falciparum 80S ribosome bound to E-tRNA | | 分子名称: | 18S ribosomal RNA, 28S ribosomal RNA, 40S ribosomal protein eS1, ... | | 著者 | Sun, M, Li, W, Blomqvist, K, Das, S, Hashem, Y, Dvorin, J.D, Frank, J. | | 登録日 | 2015-09-16 | | 公開日 | 2015-10-14 | | 最終更新日 | 2024-02-21 | | 実験手法 | ELECTRON MICROSCOPY (6.7 Å) | | 主引用文献 | Dynamical features of the Plasmodium falciparum ribosome during translation.
Nucleic Acids Res., 43, 2015
|
|
3JBN
 
 | | Cryo-electron microscopy reconstruction of the Plasmodium falciparum 80S ribosome bound to P-tRNA | | 分子名称: | 18S ribosomal RNA, 28S ribosomal RNA, 40S ribosomal protein eS1, ... | | 著者 | Sun, M, Li, W, Blomqvist, K, Das, S, Hashem, Y, Dvorin, J.D, Frank, J. | | 登録日 | 2015-09-16 | | 公開日 | 2015-10-14 | | 最終更新日 | 2024-02-21 | | 実験手法 | ELECTRON MICROSCOPY (4.7 Å) | | 主引用文献 | Dynamical features of the Plasmodium falciparum ribosome during translation.
Nucleic Acids Res., 43, 2015
|
|
5T5H
 
 | | Structure and assembly model for the Trypanosoma cruzi 60S ribosomal subunit | | 分子名称: | 40S ribosomal protein L14, 5.8S rRNA, 5S rRNA, ... | | 著者 | Liu, Z, Gutierrez-Vargas, C, Wei, J, Grassucci, R.A, Ramesh, M, Espina, N, Sun, M, Tutuncuoglu, B, Madison-Antenucci, S, Woolford Jr, J.L, Tong, L, Frank, J. | | 登録日 | 2016-08-31 | | 公開日 | 2016-10-12 | | 最終更新日 | 2019-12-18 | | 実験手法 | ELECTRON MICROSCOPY (2.54 Å) | | 主引用文献 | Structure and assembly model for the Trypanosoma cruzi 60S ribosomal subunit.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
1ZN0
 
 | | Coordinates of RRF and EF-G fitted into Cryo-EM map of the 50S subunit bound with both EF-G (GDPNP) and RRF | | 分子名称: | 16S RIBOSOMAL RNA, ELONGATION FACTOR G, Ribosome recycling factor | | 著者 | Gao, N, Zavialov, A.V, Li, W, Sengupta, J, Valle, M, Gursky, R.P, Ehrenberg, M, Frank, J. | | 登録日 | 2005-05-11 | | 公開日 | 2005-06-14 | | 最終更新日 | 2024-02-14 | | 実験手法 | ELECTRON MICROSCOPY (15.5 Å) | | 主引用文献 | Mechanism for the disassembly of the posttermination complex inferred from cryo-EM studies.
Mol.Cell, 18, 2005
|
|
1MVR
 
 | | Decoding Center & Peptidyl transferase center from the X-ray structure of the Thermus thermophilus 70S ribosome, aligned to the low resolution Cryo-EM map of E.coli 70S Ribosome | | 分子名称: | 30S RIBOSOMAL PROTEIN S12, 50S ribosomal protein L11, Helix 34 of 16S rRNA, ... | | 著者 | Rawat, U.B, Zavialov, A.V, Sengupta, J, Valle, M, Grassucci, R.A, Linde, J, Vestergaard, B, Ehrenberg, M, Frank, J. | | 登録日 | 2002-09-26 | | 公開日 | 2003-04-01 | | 最終更新日 | 2024-02-14 | | 実験手法 | ELECTRON MICROSCOPY (12.8 Å) | | 主引用文献 | A cryo-electron microscopic study of ribosome-bound termination factor RF2
Nature, 421, 2003
|
|
5TAS
 
 | | Structure of rabbit RyR1 (Caffeine/ATP/EGTA dataset, class 1) | | 分子名称: | ADENOSINE-5'-TRIPHOSPHATE, CAFFEINE, Peptidyl-prolyl cis-trans isomerase FKBP1B, ... | | 著者 | Clarke, O.B, des Georges, A, Zalk, R, Marks, A.R, Hendrickson, W.A, Frank, J. | | 登録日 | 2016-09-10 | | 公開日 | 2016-10-12 | | 最終更新日 | 2018-07-18 | | 実験手法 | ELECTRON MICROSCOPY (6.2 Å) | | 主引用文献 | Structural Basis for Gating and Activation of RyR1.
Cell, 167, 2016
|
|
5TA3
 
 | | Structure of rabbit RyR1 (Caffeine/ATP/Ca2+ dataset, class 2) | | 分子名称: | ADENOSINE-5'-TRIPHOSPHATE, CAFFEINE, CALCIUM ION, ... | | 著者 | Clarke, O.B, des Georges, A, Zalk, R, Marks, A.R, Hendrickson, W.A, Frank, J. | | 登録日 | 2016-09-09 | | 公開日 | 2016-10-12 | | 最終更新日 | 2018-07-18 | | 実験手法 | ELECTRON MICROSCOPY (4.4 Å) | | 主引用文献 | Structural Basis for Gating and Activation of RyR1.
Cell, 167, 2016
|
|
5TB2
 
 | | Structure of rabbit RyR1 (EGTA-only dataset, class 2) | | 分子名称: | Peptidyl-prolyl cis-trans isomerase FKBP1B, Ryanodine receptor 1, ZINC ION | | 著者 | Clarke, O.B, des Georges, A, Zalk, R, Marks, A.R, Hendrickson, W.A, Frank, J. | | 登録日 | 2016-09-11 | | 公開日 | 2016-10-12 | | 最終更新日 | 2024-03-13 | | 実験手法 | ELECTRON MICROSCOPY (4.6 Å) | | 主引用文献 | Structural Basis for Gating and Activation of RyR1.
Cell, 167, 2016
|
|
5TAN
 
 | | Structure of rabbit RyR1 (Caffeine/ATP/Ca2+ dataset, class 3) | | 分子名称: | ADENOSINE-5'-TRIPHOSPHATE, CAFFEINE, CALCIUM ION, ... | | 著者 | Clarke, O.B, des Georges, A, Zalk, R, Marks, A.R, Hendrickson, W.A, Frank, J. | | 登録日 | 2016-09-10 | | 公開日 | 2016-10-12 | | 最終更新日 | 2024-03-13 | | 実験手法 | ELECTRON MICROSCOPY (4.3 Å) | | 主引用文献 | Structural Basis for Gating and Activation of RyR1.
Cell, 167, 2016
|
|
5TB3
 
 | | Structure of rabbit RyR1 (EGTA-only dataset, class 3) | | 分子名称: | Peptidyl-prolyl cis-trans isomerase FKBP1B, Ryanodine receptor 1, ZINC ION | | 著者 | Clarke, O.B, des Georges, A, Zalk, R, Marks, A.R, Hendrickson, W.A, Frank, J. | | 登録日 | 2016-09-11 | | 公開日 | 2016-10-12 | | 最終更新日 | 2018-07-18 | | 実験手法 | ELECTRON MICROSCOPY (4.7 Å) | | 主引用文献 | Structural Basis for Gating and Activation of RyR1.
Cell, 167, 2016
|
|
5T9N
 
 | | Structure of rabbit RyR1 (Ca2+-only dataset, class 2) | | 分子名称: | CALCIUM ION, Peptidyl-prolyl cis-trans isomerase FKBP1B, Ryanodine receptor 1, ... | | 著者 | Clarke, O.B, des Georges, A, Zalk, R, Marks, A.R, Hendrickson, W.A, Frank, J. | | 登録日 | 2016-09-09 | | 公開日 | 2016-10-12 | | 最終更新日 | 2024-03-13 | | 実験手法 | ELECTRON MICROSCOPY (3.8 Å) | | 主引用文献 | Structural Basis for Gating and Activation of RyR1.
Cell, 167, 2016
|
|
5TAZ
 
 | | Structure of rabbit RyR1 (ryanodine dataset, class 3) | | 分子名称: | CALCIUM ION, Peptidyl-prolyl cis-trans isomerase FKBP1B, Ryanodine receptor 1, ... | | 著者 | Clarke, O.B, des Georges, A, Zalk, R, Marks, A.R, Hendrickson, W.A, Frank, J. | | 登録日 | 2016-09-10 | | 公開日 | 2016-10-12 | | 最終更新日 | 2018-07-18 | | 実験手法 | ELECTRON MICROSCOPY (4.3 Å) | | 主引用文献 | Structural Basis for Gating and Activation of RyR1.
Cell, 167, 2016
|
|
5TAV
 
 | | Structure of rabbit RyR1 (Caffeine/ATP/EGTA dataset, class 4) | | 分子名称: | ADENOSINE-5'-TRIPHOSPHATE, CAFFEINE, Peptidyl-prolyl cis-trans isomerase FKBP1B, ... | | 著者 | Clarke, O.B, des Georges, A, Zalk, R, Marks, A.R, Hendrickson, W.A, Frank, J. | | 登録日 | 2016-09-10 | | 公開日 | 2016-10-12 | | 最終更新日 | 2018-07-18 | | 実験手法 | ELECTRON MICROSCOPY (4.8 Å) | | 主引用文献 | Structural Basis for Gating and Activation of RyR1.
Cell, 167, 2016
|
|
5TAL
 
 | | Structure of rabbit RyR1 (Caffeine/ATP/Ca2+ dataset, class 1&2) | | 分子名称: | ADENOSINE-5'-TRIPHOSPHATE, CAFFEINE, CALCIUM ION, ... | | 著者 | Clarke, O.B, des Georges, A, Zalk, R, Marks, A.R, Hendrickson, W.A, Frank, J. | | 登録日 | 2016-09-10 | | 公開日 | 2016-10-12 | | 最終更新日 | 2024-03-13 | | 実験手法 | ELECTRON MICROSCOPY (4.3 Å) | | 主引用文献 | Structural Basis for Gating and Activation of RyR1.
Cell, 167, 2016
|
|
5TAW
 
 | | Structure of rabbit RyR1 (ryanodine dataset, all particles) | | 分子名称: | CALCIUM ION, Peptidyl-prolyl cis-trans isomerase FKBP1B, Ryanodine receptor 1, ... | | 著者 | Clarke, O.B, des Georges, A, Zalk, R, Marks, A.R, Hendrickson, W.A, Frank, J. | | 登録日 | 2016-09-10 | | 公開日 | 2016-10-12 | | 最終更新日 | 2018-07-18 | | 実験手法 | ELECTRON MICROSCOPY (4.4 Å) | | 主引用文献 | Structural Basis for Gating and Activation of RyR1.
Cell, 167, 2016
|
|
