3EA5
 
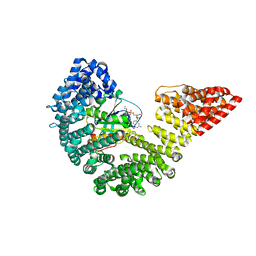 | | Kap95p Binding Induces the Switch Loops of RanGDP to adopt the GTP-bound Conformation: Implications for Nuclear Import Complex Assembly Dynamics | | Descriptor: | GTP-binding nuclear protein Ran, GUANOSINE-5'-DIPHOSPHATE, Importin subunit beta-1, ... | | Authors: | Forwood, J.K, Lonhienne, J.K, Guncar, G, Stewart, M, Marfori, M, Kobe, B. | | Deposit date: | 2008-08-24 | | Release date: | 2008-10-21 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Kap95p binding induces the switch loops of RanGDP to adopt the GTP-bound conformation: implications for nuclear import complex assembly dynamics.
J.Mol.Biol., 383, 2008
|
|
3SPS
 
 | |
2V1O
 
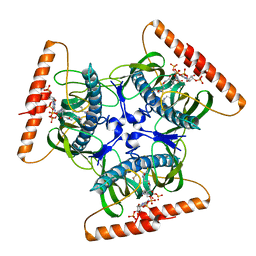 | | Crystal structure of N-terminal domain of acyl-CoA thioesterase 7 | | Descriptor: | COENZYME A, CYTOSOLIC ACYL COENZYME A THIOESTER HYDROLASE | | Authors: | Forwood, J.K, Thakur, A.S, Guncar, G, Marfori, M, Mouradov, D, Meng, W.N, Robinson, J, Huber, T, Kellie, S, Martin, J.L, Hume, D.A, Kobe, B. | | Deposit date: | 2007-05-28 | | Release date: | 2007-06-26 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Structural Basis for Recruitment of Tandem Hotdog Domains in Acyl-Coa Thioesterase 7 and its Role in Inflammation.
Proc.Natl.Acad.Sci.USA, 104, 2007
|
|
6UDS
 
 | |
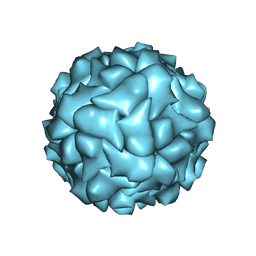
6RPK
 
 | |
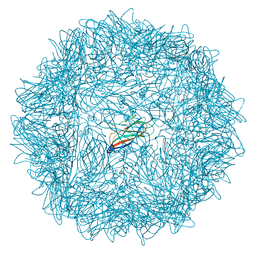
6RPO
 
 | |
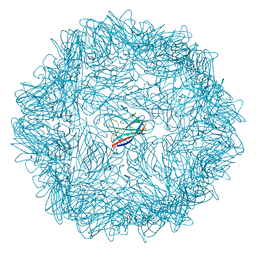
6RPL
 
 | |
7JVO
 
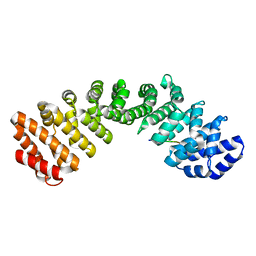 | |
3ND2
 
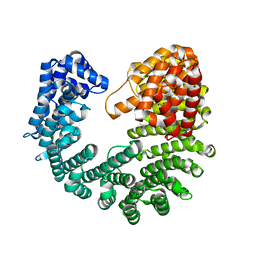 | | Structure of Yeast Importin-beta (Kap95p) | | Descriptor: | Importin subunit beta-1 | | Authors: | Forwood, J.K, Kobe, B. | | Deposit date: | 2010-06-06 | | Release date: | 2010-07-07 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Quantitative Structural Analysis of Importin-beta Flexibility: Paradigm for Solenoid Protein Structures
Structure, 18, 2010
|
|
5J37
 
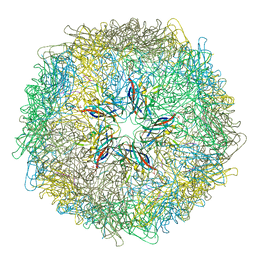 | | Crystal structure of 60-mer BFDV Capsid Protein in complex with single stranded DNA | | Descriptor: | Beak and feather disease virus capsid protein, PHOSPHATE ION, single stranded DNA | | Authors: | Sarker, S, Raidal, S, Aragao, D, Forwood, J.K. | | Deposit date: | 2016-03-30 | | Release date: | 2016-05-04 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural insights into the assembly and regulation of distinct viral capsid complexes.
Nat Commun, 7, 2016
|
|
5BYU
 
 | |
8QXW
 
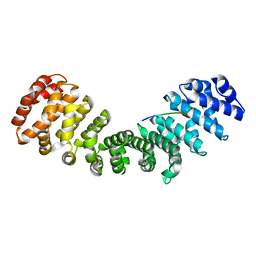 | | HCMV DNA polymerase processivity factor UL44 unphosphorylated NLS 410-433 bound to mouse importin alpha 2 | | Descriptor: | DNA polymerase processivity factor, Importin subunit alpha-1 | | Authors: | Cross, E.M, Marin, O, Ariawan, D, Aragao, D, Cozza, G, Di Iorio, E, Forwood, J.K, Alvisi, G. | | Deposit date: | 2023-10-25 | | Release date: | 2023-11-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural determinants of phosphorylation-dependent nuclear transport of HCMV DNA polymerase processivity factor UL44.
Febs Lett., 598, 2024
|
|
8QXX
 
 | | HCMV DNA polymerase processivity factor UL44 phosphorylated NLS 410-433 bound to mouse importin alpha 2 | | Descriptor: | DNA polymerase processivity factor, Importin subunit alpha-1 | | Authors: | Cross, E.M, Marin, O, Ariawan, D, Aragao, D, Cozza, G, Di Iorio, E, Forwood, J.K, Alvisi, G. | | Deposit date: | 2023-10-25 | | Release date: | 2023-11-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural determinants of phosphorylation-dependent nuclear transport of HCMV DNA polymerase processivity factor UL44.
Febs Lett., 598, 2024
|
|
8Q8K
 
 | | KI Polyomavirus LTA NLS bound to importin alpha 2 | | Descriptor: | Importin subunit alpha-1, Large T antigen | | Authors: | Cross, E.M, Forwood, J.K, Alvisi, G. | | Deposit date: | 2023-08-18 | | Release date: | 2024-02-07 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | A functional and structural comparative analysis of large tumor antigens reveals evolution of different importin alpha-dependent nuclear localization signals.
Protein Sci., 33, 2024
|
|
6WPR
 
 | |
8VPU
 
 | |
8G8R
 
 | |
8G8S
 
 | |
8G8Q
 
 | |
7N8J
 
 | |
7RG6
 
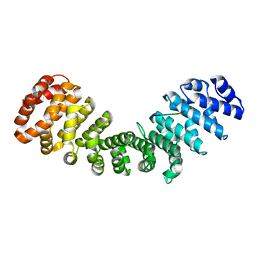 | | Importin alpha2 in complex with ORF4B Bat coronavirus HKU5 | | Descriptor: | Importin subunit alpha-1, Non-structural protein ORF4b | | Authors: | Foster, J.K, Tsimbalyuk, S, Roby, J.A, Aragao, D, Forwood, J.K. | | Deposit date: | 2021-07-14 | | Release date: | 2021-08-25 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | MERS-CoV ORF4b employs an unusual binding mechanism to target IMP alpha and block innate immunity.
Nat Commun, 13, 2022
|
|
6WX9
 
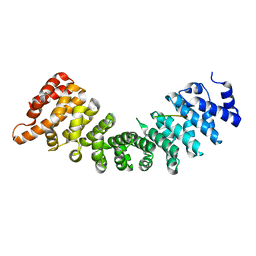 | | SOX2 bound to Importin-alpha 5 | | Descriptor: | Importin subunit alpha-5, Transcription factor SOX-2 | | Authors: | Bikshapathi, J, Stewart, M, Forwood, J.K, Aragao, D, Roman, N. | | Deposit date: | 2020-05-10 | | Release date: | 2020-10-28 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis for nuclear import selectivity of pioneer transcription factor SOX2.
Nat Commun, 12, 2021
|
|
6WX8
 
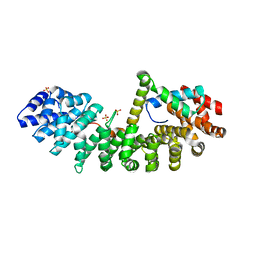 | | SOX2 bound to Importin-alpha 3 | | Descriptor: | Importin subunit alpha-3, SULFATE ION, Transcription factor SOX-2 | | Authors: | Bikshapathi, J, Stewart, M, Forwood, J.K, Aragao, D, Roman, N. | | Deposit date: | 2020-05-10 | | Release date: | 2020-10-28 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for nuclear import selectivity of pioneer transcription factor SOX2.
Nat Commun, 12, 2021
|
|
5KLT
 
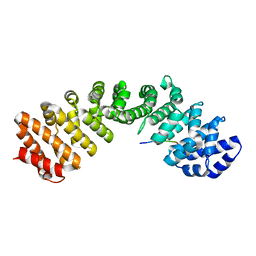 | | Prototypical P4[M]cNLS | | Descriptor: | Importin subunit alpha-1, Prototypical P4[M]cNLS | | Authors: | Smith, K.M, Forwood, J.K. | | Deposit date: | 2016-06-25 | | Release date: | 2016-07-27 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Contribution of the residue at position 4 within classical nuclear localization signals to modulating interaction with importins and nuclear targeting.
Biochim Biophys Acta Mol Cell Res, 1865, 2018
|
|
5KLR
 
 | | Prototypical P4[R]cNLS | | Descriptor: | Importin subunit alpha-1, Prototypical P4[R]cNLS | | Authors: | Smith, K.M, Forwood, J.K. | | Deposit date: | 2016-06-25 | | Release date: | 2016-07-27 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Contribution of the residue at position 4 within classical nuclear localization signals to modulating interaction with importins and nuclear targeting.
Biochim Biophys Acta Mol Cell Res, 1865, 2018
|
|
