1PTX
 
 | |
1RCN
 
 | | CRYSTAL STRUCTURE OF THE RIBONUCLEASE A D(APTPAPAPG) COMPLEX : DIRECT EVIDENCE FOR EXTENDED SUBSTRATE RECOGNITION | | Descriptor: | DNA (5'-D(*AP*TP*AP*A)-3'), PROTEIN (RIBONUCLEASE A (E.C.3.1.27.5)) | | Authors: | Fontecilla-Camps, J.C, De Llorens, R, Le Du, M.H, Cuchillo, C.M. | | Deposit date: | 1994-05-27 | | Release date: | 1994-09-30 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Crystal structure of ribonuclease A.d(ApTpApApG) complex. Direct evidence for extended substrate recognition.
J.Biol.Chem., 269, 1994
|
|
4URH
 
 | | High-resolution structure of partially oxidized D. fructosovorans NiFe-hydrogenase | | Descriptor: | CARBONMONOXIDE-(DICYANO) IRON, FE3-S4 CLUSTER, GLYCEROL, ... | | Authors: | Volbeda, A, Martin, L, Barbier, E, Gutierrez-Sanz, O, DeLacey, A.L, Liebgott, P.P, Dementin, S, Rousset, M, Fontecilla-Camps, J.C. | | Deposit date: | 2014-06-30 | | Release date: | 2014-10-29 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | Crystallographic studies of [NiFe]-hydrogenase mutants: towards consensus structures for the elusive unready oxidized states.
J. Biol. Inorg. Chem., 20, 2015
|
|
3USC
 
 | | Crystal Structure of E. coli hydrogenase-1 in a ferricyanide-oxidized form | | Descriptor: | CARBONMONOXIDE-(DICYANO) IRON, CHLORIDE ION, DODECYL-BETA-D-MALTOSIDE, ... | | Authors: | Volbeda, A, Fontecilla-Camps, J.C, Darnault, C. | | Deposit date: | 2011-11-23 | | Release date: | 2012-03-28 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | X-ray crystallographic and computational studies of the O2-tolerant [NiFe]-hydrogenase 1 from Escherichia coli.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
1MD7
 
 | | Monomeric structure of the zymogen of complement protease C1r | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, C1R COMPLEMENT SERINE PROTEASE | | Authors: | Budayova-Spano, M, Grabarse, W, Thielens, N.M, Hillen, H, Lacroix, M, Schmidt, M, Fontecilla-Camps, J, Arlaud, G.J, Gaboriaud, C. | | Deposit date: | 2002-08-07 | | Release date: | 2003-08-07 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Monomeric structures of the zymogen and active catalytic domain of complement protease c1r: further insights into the c1 activation mechanism
Structure, 10, 2002
|
|
1MD8
 
 | | Monomeric structure of the active catalytic domain of complement protease C1r | | Descriptor: | C1R COMPLEMENT SERINE PROTEASE | | Authors: | Budayova-Spano, M, Grabarse, W, Thielens, N.M, Hillen, H, Lacroix, M, Schmidt, M, Fontecilla-Camps, J, Arlaud, G.J, Gaboriaud, C. | | Deposit date: | 2002-08-07 | | Release date: | 2003-08-07 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Monomeric structures of the zymogen and active catalytic domain of complement protease c1r: further insights into the c1 activation mechanism
Structure, 10, 2002
|
|
5CVR
 
 | |
7B0C
 
 | | [4Fe-4S]-NsrR complexed to 23-bp HmpA1 operator fragment | | Descriptor: | DNA (5'-D(P*AP*AP*CP*AP*CP*GP*AP*AP*TP*AP*TP*CP*AP*TP*CP*TP*AP*CP*CP*AP*AP*TP*T)-3'), DNA (5'-D(P*AP*AP*TP*TP*GP*GP*TP*AP*GP*AP*TP*GP*AP*TP*AP*TP*TP*CP*GP*TP*GP*TP*T)-3'), HTH-type transcriptional repressor NsrR, ... | | Authors: | Rohac, R, Fontecilla-Camps, J.C, Volbeda, A. | | Deposit date: | 2020-11-19 | | Release date: | 2022-06-01 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural determinants of DNA recognition by the NO sensor NsrR and related Rrf2-type [FeS]-transcription factors.
Commun Biol, 5, 2022
|
|
8QTO
 
 | | CRYSTAL STRUCTURE OF HOLO-L28H-FNR OF A. FISCHERI | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, FNR type regulator, IRON/SULFUR CLUSTER | | Authors: | Volbeda, A, Fontecilla-Camps, J.C. | | Deposit date: | 2023-10-13 | | Release date: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Probing the Reactivity of [4Fe-4S] Fumarate and Nitrate Reduction (FNR) Regulator with O2 and NO: Increased O2 Resistance and Relative Specificity for NO of the [4Fe-4S] L28H FNR Cluster
Inorganics (Basel), 11, 2023
|
|
6Y45
 
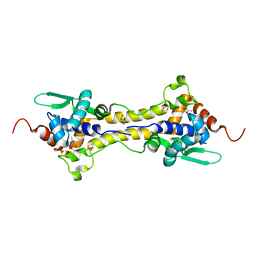 | | Crystal Structure of the H33A variant of RsrR | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Rohac, R, Volbeda, A, Fontecilla-Camps, J.C. | | Deposit date: | 2020-02-19 | | Release date: | 2020-03-04 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Electron and Proton Transfers Modulate DNA Binding by the Transcription Regulator RsrR.
J.Am.Chem.Soc., 142, 2020
|
|
6Y42
 
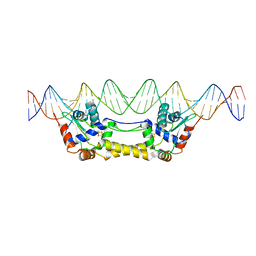 | |
4UQP
 
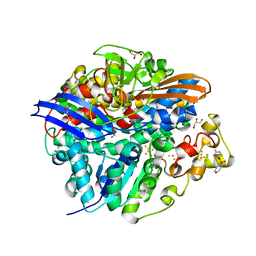 | | High-resolution structure of the D. fructosovorans NiFe-hydrogenase L122A mutant after exposure to air | | Descriptor: | CARBONMONOXIDE-(DICYANO) IRON, FE3-S4 CLUSTER, GLYCEROL, ... | | Authors: | Volbeda, A, Martin, L, Barbier, E, Gutierrez-Sanz, O, DeLacey, A.L, Liebgott, P.P, Dementin, S, Rousset, M, Fontecilla-Camps, J.C. | | Deposit date: | 2014-06-24 | | Release date: | 2014-10-29 | | Last modified: | 2023-03-15 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Crystallographic Studies of [Nife]-Hydrogenase Mutants: Towards Consensus Structures for the Elusive Unready Oxidized States.
J.Biol.Inorg.Chem., 20, 2015
|
|
4UQL
 
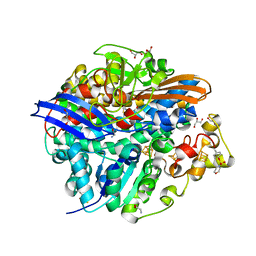 | | High-resolution structure of a Ni-A Ni-Sox mixture of the D. fructosovorans NiFe-hydrogenase L122A mutant | | Descriptor: | CARBONMONOXIDE-(DICYANO) IRON, CHLORIDE ION, FE3-S4 CLUSTER, ... | | Authors: | Volbeda, A, Martin, L, Barbier, E, Gutierrez-Sanz, O, DeLacey, A.L, Liebgott, P.P, Dementin, S, Rousset, M, Fontecilla-Camps, J.C. | | Deposit date: | 2014-06-24 | | Release date: | 2014-10-29 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.22 Å) | | Cite: | Crystallographic Studies of [Nife]-Hydrogenase Mutants: Towards Consensus Structures for the Elusive Unready Oxidized States.
J.Biol.Inorg.Chem., 20, 2015
|
|
3CIW
 
 | | X-RAY structure of the [FeFe]-hydrogenase maturase HydE from thermotoga maritima | | Descriptor: | 3-[(3-CHOLAMIDOPROPYL)DIMETHYLAMMONIO]-1-PROPANESULFONATE, CHLORIDE ION, FeFe-Hydrogenase maturase, ... | | Authors: | Nicolet, Y, Ruback, J.K, Posewitz, M.C, Amara, P, Mathevon, C, Atta, M, Fontecave, M, Fontecilla-Camps, J.C. | | Deposit date: | 2008-03-12 | | Release date: | 2008-04-08 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | X-ray Structure of the [FeFe]-Hydrogenase Maturase HydE from Thermotoga maritima
J.Biol.Chem., 283, 2008
|
|
6G94
 
 | |
4UPV
 
 | | Low X-ray dose structure of a Ni-A Ni-Sox mixture of the D. fructosovorans NiFe-hydrogenase L122A mutant | | Descriptor: | CARBONMONOXIDE-(DICYANO) IRON, CHLORIDE ION, FE3-S4 CLUSTER, ... | | Authors: | Volbeda, A, Martin, L, Barbier, E, Gutierrez-Sanz, O, DeLacey, A.L, Liebgott, P.P, Dementin, S, Rousset, M, Fontecilla-Camps, J.C. | | Deposit date: | 2014-06-18 | | Release date: | 2014-10-29 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Crystallographic studies of [NiFe]-hydrogenase mutants: towards consensus structures for the elusive unready oxidized states.
J. Biol. Inorg. Chem., 20, 2015
|
|
4UPE
 
 | | Structure of the unready Ni-A state of the S499C mutant of D. fructosovorans NiFe-hydrogenase | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, CARBONMONOXIDE-(DICYANO) IRON, ... | | Authors: | Volbeda, A, Martin, L, Barbier, E, Gutierrez-Sanz, O, DeLacey, A.L, Liebgott, P.P, Dementin, S, Rousset, M, Fontecilla-Camps, J.C. | | Deposit date: | 2014-06-16 | | Release date: | 2014-10-29 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystallographic studies of [NiFe]-hydrogenase mutants: towards consensus structures for the elusive unready oxidized states.
J. Biol. Inorg. Chem., 20, 2015
|
|
3IIZ
 
 | | X-ray structure of the FeFe-hydrogenase maturase HydE from T. maritima in complex with S-adenosyl-L-methionine | | Descriptor: | 3-[(3-CHOLAMIDOPROPYL)DIMETHYLAMMONIO]-1-PROPANESULFONATE, Biotin synthetase, putative, ... | | Authors: | Nicolet, Y, Amara, P, Mouesca, J.M, Fontecilla-Camps, J.C. | | Deposit date: | 2009-08-03 | | Release date: | 2009-09-22 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Unexpected electron transfer mechanism upon AdoMet cleavage in radical SAM proteins
Proc.Natl.Acad.Sci.USA, 2009
|
|
3IIX
 
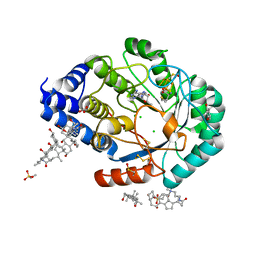 | | X-ray structure of the FeFe-hydrogenase maturase HydE from T. maritima in complex with methionine and 5'deoxyadenosine | | Descriptor: | 3-[(3-CHOLAMIDOPROPYL)DIMETHYLAMMONIO]-1-PROPANESULFONATE, 5'-DEOXYADENOSINE, CARBONATE ION, ... | | Authors: | Nicolet, Y, Amara, P, Mouesca, J.M, Fontecilla-Camps, J.C. | | Deposit date: | 2009-08-03 | | Release date: | 2009-09-22 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Unexpected electron transfer mechanism upon AdoMet cleavage in radical SAM proteins
Proc.Natl.Acad.Sci.USA, 2009
|
|
3UQY
 
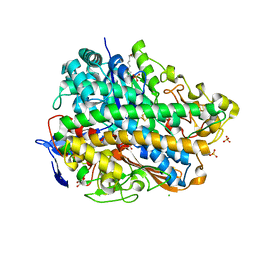 | | H2-reduced structure of E. coli hydrogenase-1 | | Descriptor: | CARBONMONOXIDE-(DICYANO) IRON, CHLORIDE ION, DODECYL-BETA-D-MALTOSIDE, ... | | Authors: | Volbeda, A, Fontecilla-Camps, J.C, Darnault, C. | | Deposit date: | 2011-11-21 | | Release date: | 2012-03-28 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | X-ray crystallographic and computational studies of the O2-tolerant [NiFe]-hydrogenase 1 from Escherichia coli.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3USE
 
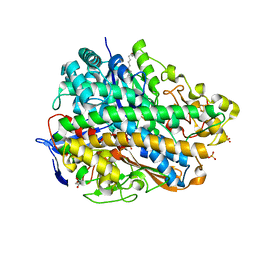 | | Crystal Structure of E. coli hydrogenase-1 in its as-isolated form | | Descriptor: | CARBONMONOXIDE-(DICYANO) IRON, CHLORIDE ION, DODECYL-BETA-D-MALTOSIDE, ... | | Authors: | Volbeda, A, Fontecilla-Camps, J.C, Darnault, C. | | Deposit date: | 2011-11-23 | | Release date: | 2012-03-28 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | X-ray crystallographic and computational studies of the O2-tolerant [NiFe]-hydrogenase 1 from Escherichia coli.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
5LQS
 
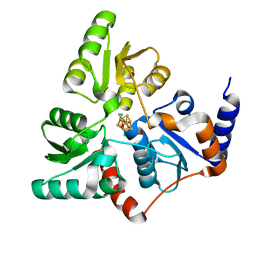 | | Structure of quinolinate synthase Y21F mutant in complex with substrate-derived quinolinate | | Descriptor: | CHLORIDE ION, IRON/SULFUR CLUSTER, QUINOLINIC ACID, ... | | Authors: | Volbeda, A, Fontecilla-Camps, J.C. | | Deposit date: | 2016-08-17 | | Release date: | 2016-08-31 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structures of Quinolinate Synthase in Complex with a Substrate Analogue, the Condensation Intermediate, and Substrate-Derived Product.
J.Am.Chem.Soc., 138, 2016
|
|
6F48
 
 | | Structure of quinolinate synthase with reaction intermediates X and Y | | Descriptor: | 2-imino,3-carboxy,5-hydroxy,6-oxo hexanoic acid, 5-hydroxy,-4,5-dihydroquinolinate, CHLORIDE ION, ... | | Authors: | Volbeda, A, Fontecilla-Camps, J.C. | | Deposit date: | 2017-11-29 | | Release date: | 2018-04-25 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystallographic Trapping of Reaction Intermediates in Quinolinic Acid Synthesis by NadA.
ACS Chem. Biol., 13, 2018
|
|
6F4L
 
 | |
6F4D
 
 | |
