4IMF
 
 | |
4IMD
 
 | |
4IMC
 
 | |
4IME
 
 | |
2II6
 
 | |
2NN3
 
 | | structure of pro-sf-caspase-1 | | Descriptor: | Caspase-1 | | Authors: | Fisher, A.J, Ni, L. | | Deposit date: | 2006-10-23 | | Release date: | 2007-10-23 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Pro-sfcapsase-1, structural insights into activation mechanism of caspases
To be Published
|
|
5ED1
 
 | | Human Adenosine Deaminase Acting on dsRNA (ADAR2) mutant E488Q bound to dsRNA sequence derived from S. cerevisiae BDF2 gene | | Descriptor: | Double-stranded RNA-specific editase 1, INOSITOL HEXAKISPHOSPHATE, RNA (5'-R(*GP*AP*CP*UP*GP*AP*AP*CP*GP*AP*CP*CP*AP*AP*UP*GP*UP*GP*GP*GP*GP*AP*A)-3'), ... | | Authors: | Matthews, M.M, Fisher, A.J, Beal, P.A. | | Deposit date: | 2015-10-20 | | Release date: | 2016-04-13 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.77 Å) | | Cite: | Structures of human ADAR2 bound to dsRNA reveal base-flipping mechanism and basis for site selectivity.
Nat.Struct.Mol.Biol., 23, 2016
|
|
2G18
 
 | |
1M7G
 
 | | Crystal structure of APS kinase from Penicillium Chrysogenum: Ternary structure with ADP and APS | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-DIPHOSPHATE-2',3'-VANADATE, ADENOSINE-5'-PHOSPHOSULFATE, ... | | Authors: | Lansdon, E.B, Segel, I.H, Fisher, A.J. | | Deposit date: | 2002-07-19 | | Release date: | 2002-11-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Ligand-Induced Structural Changes in Adenosine 5'-Phosphosulfate
Kinase from Penicillium chrysogenum.
Biochemistry, 41, 2002
|
|
1M7H
 
 | | Crystal Structure of APS kinase from Penicillium Chrysogenum: Structure with APS soaked out of one dimer | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-PHOSPHOSULFATE, Adenylylsulfate kinase, ... | | Authors: | Lansdon, E.B, Sege, I.H, Fisher, A.J. | | Deposit date: | 2002-07-19 | | Release date: | 2002-11-27 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Ligand-Induced Structural Changes in Adenosine 5'-Phosphosulfate
Kinase from Penicillium chrysogenum.
Biochemistry, 41, 2002
|
|
1M8P
 
 | |
1M0O
 
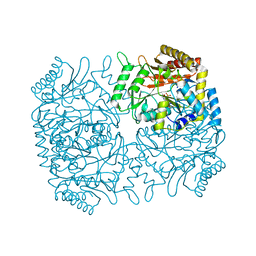 | | Structure of Dialkylglycine Decarboxylase Complexed with 1-Amino-1-methylpropanephosphonate | | Descriptor: | (1R)-1-[((1E)-{3-HYDROXY-2-METHYL-5-[(PHOSPHONOOXY)METHYL]PYRIDIN-4-YL}METHYLENE)AMINO]-1-METHYLPROPYLPHOSPHONIC ACID, 2,2-Dialkylglycine decarboxylase, POTASSIUM ION, ... | | Authors: | Liu, W, Rogers, C.J, Fisher, A.J, Toney, M.D. | | Deposit date: | 2002-06-13 | | Release date: | 2002-10-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Aminophosphonate Inhibitors of Dialkylglycine Decarboxylase: Structural Basis for Slow Binding Inhibition
Biochemistry, 41, 2002
|
|
1M72
 
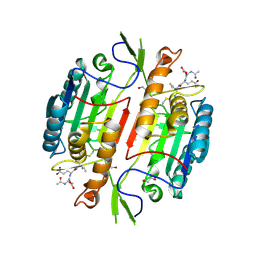 | | Crystal Structure of Caspase-1 from Spodoptera frugiperda | | Descriptor: | 1,2-ETHANEDIOL, Ace-Asp-Glu-Val-Asp-chloromethylketone, Caspase-1 | | Authors: | Forsyth, C.M, Lemongello, D, Friesen, P.D, Fisher, A.J. | | Deposit date: | 2002-07-18 | | Release date: | 2004-01-20 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of an invertebrate caspase.
J.Biol.Chem., 279, 2004
|
|
8E0F
 
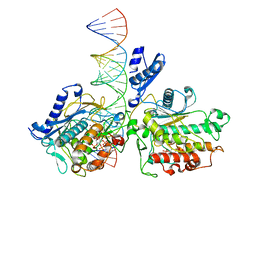 | | Human Adenosine Deaminase Acting on dsRNA (ADAR2-RD) bound to dsRNA containing a G-G pair adjacent to the target site | | Descriptor: | Double-stranded RNA-specific editase 1, INOSITOL HEXAKISPHOSPHATE, RNA (5-R(*GP*CP*UP*CP*GP*CP*GP*AP*UP*GP*CP*GP*(8AZ)P*GP*AP*GP*GP*GP*CP* UP*CP*UP*GP*AP*UP*AP*GP*CP*UP*AP*CP*G)-3), ... | | Authors: | Wilcox, X.E, Fisher, A.J, Beal, P.A. | | Deposit date: | 2022-08-09 | | Release date: | 2022-10-26 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | ADAR activation by inducing a syn conformation at guanosine adjacent to an editing site.
Nucleic Acids Res., 50, 2022
|
|
5XEP
 
 | | Crystal structure of BRP39, a chitinase-like protein, at 2.6 Angstorm resolution | | Descriptor: | 1,2-ETHANEDIOL, Chitinase-3-like protein 1 | | Authors: | Mohanty, A.K, Fisher, A.J, Choudhary, S, Kaushik, J.K. | | Deposit date: | 2017-04-05 | | Release date: | 2018-04-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structure of BRP39, a signalling glycoprotein expressed during mammary gland apoptosis.
To be published
|
|
6VFF
 
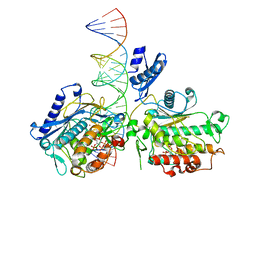 | | Dimer of Human Adenosine Deaminase Acting on dsRNA (ADAR2) mutant E488Q bound to dsRNA sequence derived from human GLI1 gene | | Descriptor: | Double-stranded RNA-specific editase 1, INOSITOL HEXAKISPHOSPHATE, RNA (5-R(*GP*CP*UP*CP*GP*CP*GP*AP*UP*GP*CP*UP*(8AZ)P*GP*AP*GP*GP*GP*CP* UP*CP*UP*GP*AP*UP*AP*GP*CP*UP*AP*CP*G)-3), ... | | Authors: | Thuy-boun, A.S, Fisher, A.J, Beal, P.A. | | Deposit date: | 2020-01-03 | | Release date: | 2020-07-01 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Asymmetric dimerization of adenosine deaminase acting on RNA facilitates substrate recognition.
Nucleic Acids Res., 48, 2020
|
|
5HP2
 
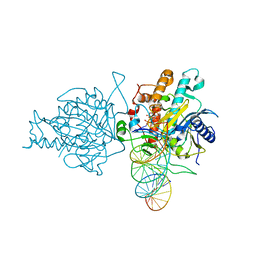 | | Human Adenosine Deaminase Acting on dsRNA (ADAR2) bound to dsRNA sequence derived from S. cerevisiae BDF2 gene with AU basepair at reaction site | | Descriptor: | Double-stranded RNA-specific editase 1, INOSITOL HEXAKISPHOSPHATE, RNA (5'-R(*GP*AP*CP*UP*GP*AP*AP*CP*GP*AP*CP*UP*AP*AP*UP*GP*UP*GP*GP*GP*GP*AP*A)-3'), ... | | Authors: | Matthews, M.M, Fisher, A.J, Beal, P.A. | | Deposit date: | 2016-01-20 | | Release date: | 2016-04-13 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.983 Å) | | Cite: | Structures of human ADAR2 bound to dsRNA reveal base-flipping mechanism and basis for site selectivity.
Nat.Struct.Mol.Biol., 23, 2016
|
|
5HP3
 
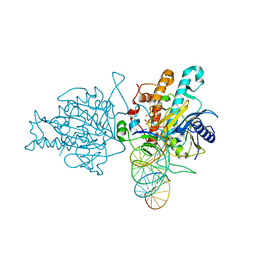 | | Human Adenosine Deaminase Acting on dsRNA (ADAR2) bound to dsRNA sequence derived from S. cerevisiae BDF2 gene with AC mismatch at reaction site | | Descriptor: | Double-stranded RNA-specific editase 1, INOSITOL HEXAKISPHOSPHATE, RNA (5'-R(*GP*AP*CP*UP*GP*AP*AP*CP*GP*AP*CP*CP*AP*AP*UP*GP*UP*GP*GP*GP*GP*AP*A)-3'), ... | | Authors: | Matthews, M.M, Fisher, A.J, Beal, P.A. | | Deposit date: | 2016-01-20 | | Release date: | 2016-04-13 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.091 Å) | | Cite: | Structures of human ADAR2 bound to dsRNA reveal base-flipping mechanism and basis for site selectivity.
Nat.Struct.Mol.Biol., 23, 2016
|
|
4R36
 
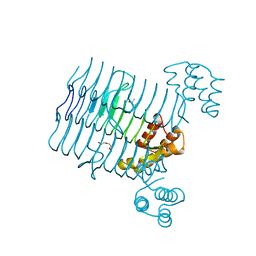 | | Crystal structure analysis of LpxA, a UDP-N-acetylglucosamine acyltransferase from Bacteroides fragilis 9343 | | Descriptor: | 2-(2-METHOXYETHOXY)ETHANOL, 3,6,9,12,15,18,21,24-OCTAOXAHEXACOSAN-1-OL, ACETATE ION, ... | | Authors: | Ngo, A, Fong, K, Cox, D, Fisher, A, Chen, X. | | Deposit date: | 2014-08-14 | | Release date: | 2015-05-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structures of Bacteroides fragilis uridine 5'-diphosphate-N-acetylglucosamine (UDP-GlcNAc) acyltransferase (BfLpxA).
Acta Crystallogr.,Sect.D, 71, 2015
|
|
7KFN
 
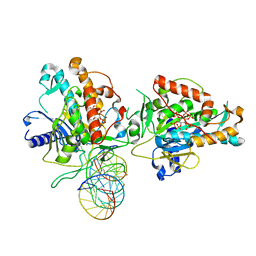 | | Structure of Human Adenosine Deaminase Acting on dsRNA (ADAR2) bound to dsRNA containing a 2'-deoxy Benner's Base Z opposite the edited base | | Descriptor: | Double-stranded RNA-specific editase 1, Gli1 1W5 23mer RNA, Gli1 8AZ 23mer RNA, ... | | Authors: | Wilcox, X.E, Fisher, A.J, Beal, P.A. | | Deposit date: | 2020-10-14 | | Release date: | 2021-06-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Rational Design of RNA Editing Guide Strands: Cytidine Analogs at the Orphan Position.
J.Am.Chem.Soc., 143, 2021
|
|
1NC2
 
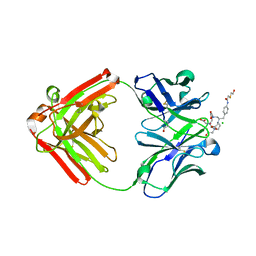 | | Crystal Structure of Monoclonal Antibody 2D12.5 Fab Complexed with Y-DOTA | | Descriptor: | (S)-2-(4-(2-(2-HYDROXYETHYLTHIO)-ACETAMIDO)-BENZYL)-1,4,7,10-TETRAAZACYCLODODECANE-N,N',N'',N'''-TETRAACETATE, 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, ... | | Authors: | Corneillie, T.M, Fisher, A.J, Meares, C.F. | | Deposit date: | 2002-12-04 | | Release date: | 2003-12-16 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structures of two complexes of the rare-earth-DOTA-binding antibody 2D12.5: ligand generality from a chiral system.
J.Am.Chem.Soc., 125, 2003
|
|
1NC4
 
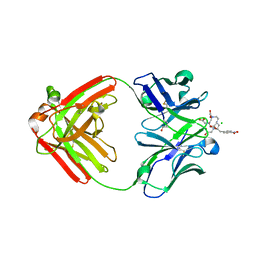 | | Crystal Structure of Monoclonal Antibody 2D12.5 Fab Complexed with Gd-DOTA | | Descriptor: | (S)-2-(4-NITROBENZYL)-1,4,7,10-TETRAAZACYCLODODECANE-N,N',N'',N'''-TETRAACETATE, 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, ... | | Authors: | Corneillie, T.M, Fisher, A.J, Meares, C.F. | | Deposit date: | 2002-12-04 | | Release date: | 2003-12-16 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal structures of two complexes of the rare-earth-DOTA-binding antibody 2D12.5: ligand generality from a chiral system.
J.Am.Chem.Soc., 125, 2003
|
|
3CS1
 
 | | Flagellar Calcium-binding Protein (FCaBP) from T. cruzi | | Descriptor: | Flagellar calcium-binding protein | | Authors: | Ames, J.B, Ladner, J.E, Wingard, J.N, Robinson, H, Fisher, A. | | Deposit date: | 2008-04-08 | | Release date: | 2008-06-24 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Insights into Membrane Targeting by the Flagellar Calcium-binding Protein (FCaBP), a Myristoylated and Palmitoylated Calcium Sensor in Trypanosoma cruzi.
J.Biol.Chem., 283, 2008
|
|
6XHG
 
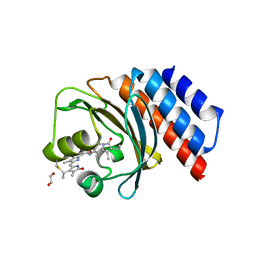 | | Far-red absorbing dark state of JSC1_58120g3 with bound biliverdin IXa (BV) | | Descriptor: | 1,2-ETHANEDIOL, 3-[2-[(~{Z})-[5-[(4-ethenyl-3-methyl-5-oxidanylidene-pyrrol-2-ylidene)methyl]-3-(3-hydroxy-3-oxopropyl)-4-methyl-pyrrol-2-ylidene]methyl]-5-[(~{Z})-(3-ethyl-4-methyl-5-oxidanylidene-pyrrol-2-ylidene)methyl]-4-methyl-1~{H}-pyrrol-3-yl]propanoic acid, JSC1_58120g3 | | Authors: | Moreno, M.V, Rockwell, N.C, Fisher, A.J, Lagarias, J.C. | | Deposit date: | 2020-06-18 | | Release date: | 2020-10-28 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A far-red cyanobacteriochrome lineage specific for verdins.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
1I3S
 
 | | THE 2.7 ANGSTROM RESOLUTION CRYSTAL STRUCTURE OF A MUTATED BACULOVIRUS P35 AFTER CASPASE CLEAVAGE | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, EARLY 35 KDA PROTEIN | | Authors: | dela Cruz, W.P, Lemongello, D, Friesen, P.D, Fisher, A.J. | | Deposit date: | 2001-02-15 | | Release date: | 2001-10-24 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of baculovirus P35 reveals a novel conformational change in the reactive site loop after caspase cleavage.
J.Biol.Chem., 276, 2001
|
|
