6D06
 
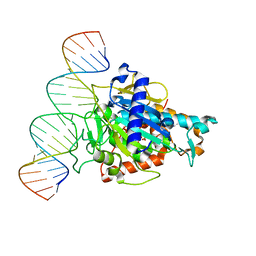 | | Human ADAR2d E488Y mutant complexed with dsRNA containing an abasic site opposite the edited base | | Descriptor: | Double-stranded RNA-specific editase 1, INOSITOL HEXAKISPHOSPHATE, RNA (5'-R(*CP*AP*GP*AP*GP*CP*CP*CP*CP*CP*NP*AP*GP*CP*AP*UP*CP*GP*CP*GP*AP*GP*C)-3'), ... | | Authors: | Matthews, M.M, Fisher, A.J, Beal, P.A. | | Deposit date: | 2018-04-10 | | Release date: | 2019-02-20 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | A Bump-Hole Approach for Directed RNA Editing.
Cell Chem Biol, 26, 2019
|
|
6BN0
 
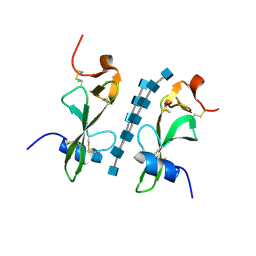 | | Avirulence protein 4 (Avr4) from Cladosporium fulvum bound to the hexasaccharide of chitin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, Race-specific elicitor A4 | | Authors: | Hurlburt, N.K, Fisher, A.J. | | Deposit date: | 2017-11-15 | | Release date: | 2018-08-22 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure of the Cladosporium fulvum Avr4 effector in complex with (GlcNAc)6 reveals the ligand-binding mechanism and uncouples its intrinsic function from recognition by the Cf-4 resistance protein.
PLoS Pathog., 14, 2018
|
|
5DJF
 
 | |
5TOS
 
 | |
3NM8
 
 | | Crystal structure of Tyrosinase from Bacillus megaterium | | Descriptor: | CHLORIDE ION, COPPER (II) ION, Tyrosinase, ... | | Authors: | Sendovski, M, Kanteev, M, Adir, N, Fishman, A. | | Deposit date: | 2010-06-22 | | Release date: | 2010-11-17 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | First structures of an active bacterial tyrosinase reveal copper plasticity
J.Mol.Biol., 405, 2011
|
|
6S3J
 
 | | Crystal Structure of lipase from Geobacillus stearothermophilus T6 variant E134C/F149C | | Descriptor: | CALCIUM ION, Lipase, ZINC ION | | Authors: | Gihaz, S, Bash, Y, Rush, I, Shahar, A, Pazy, Y, Fishman, A. | | Deposit date: | 2019-06-25 | | Release date: | 2019-10-09 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Bridges to Stability: Engineering Disulfide Bonds Towards Enhanced Lipase Biodiesel Synthesis
Chemcatchem, 2019
|
|
6EI4
 
 | | Crystal Structure of tyrosinase from Bacillus megaterium with B5N inhibitor in the active site | | Descriptor: | COPPER (II) ION, Tyrosinase, [4-[(4-fluorophenyl)methyl]piperazin-1-yl]-(2-methylphenyl)methanone | | Authors: | Deri, B, Gitto, R, Pazy Benhar, Y, Fishman, A. | | Deposit date: | 2017-09-17 | | Release date: | 2018-04-25 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Targeting Tyrosinase: Development and Structural Insights of Novel Inhibitors Bearing Arylpiperidine and Arylpiperazine Fragments.
J. Med. Chem., 61, 2018
|
|
6EM0
 
 | | Crystal Structure of 2-hydroxybiphenyl 3-monooxygenase M321A from Pseudomonas azelaica | | Descriptor: | 2-hydroxybiphenyl-3-monooxygenase, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Deri, B, Bregman-Cohen, A, Pazy Benhar, Y, Fishman, A. | | Deposit date: | 2017-10-01 | | Release date: | 2018-01-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.78 Å) | | Cite: | Altering 2-Hydroxybiphenyl 3-Monooxygenase Regioselectivity by Protein Engineering for the Production of a New Antioxidant.
Chembiochem, 19, 2018
|
|
3NQ0
 
 | | Crystal Structure of Tyrosinase from Bacillus megaterium crystallized in the absence of Zinc | | Descriptor: | COPPER (II) ION, Tyrosinase | | Authors: | Sendovski, M, Kanteev, M, Adir, N, Fishman, A. | | Deposit date: | 2010-06-29 | | Release date: | 2010-11-17 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | First structures of an active bacterial tyrosinase reveal copper plasticity.
J.Mol.Biol., 405, 2011
|
|
6CKK
 
 | |
4X71
 
 | | Crystal Structure of lipase from Geobacillus stearothermophilus T6 methanol stable variant A269T | | Descriptor: | CALCIUM ION, Lipase, ZINC ION | | Authors: | Kanteev, M, Dror, A, Gihaz, S, Shahar, A, Fishman, A. | | Deposit date: | 2014-12-09 | | Release date: | 2015-06-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural insights into methanol-stable variants of lipase T6 from Geobacillus stearothermophilus.
Appl.Microbiol.Biotechnol., 99, 2015
|
|
4Z4A
 
 | |
4Z2U
 
 | |
6S3V
 
 | | Crystal Structure of lipase from Geobacillus stearothermophilus T6 methanol stable variant E251C/G332C | | Descriptor: | CALCIUM ION, Lipase, ZINC ION | | Authors: | Gihaz, S, Bash, Y, Rush, I, Shahar, A, Pazy, Y, Fishman, A. | | Deposit date: | 2019-06-26 | | Release date: | 2019-10-09 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Bridges to Stability: Engineering Disulfide Bonds Towards Enhanced Lipase Biodiesel Synthesis
Chemcatchem, 2019
|
|
4X7B
 
 | | Crystal Structure of lipase from Geobacillus stearothermophilus T6 methanol stable variant H86Y/A269T | | Descriptor: | CALCIUM ION, Lipase, ZINC ION | | Authors: | Kanteev, M, Dror, A, Gihaz, S, Fishman, A. | | Deposit date: | 2014-12-09 | | Release date: | 2015-06-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural insights into methanol-stable variants of lipase T6 from Geobacillus stearothermophilus.
Appl.Microbiol.Biotechnol., 99, 2015
|
|
4X6U
 
 | | Crystal Structure of lipase from Geobacillus stearothermophilus T6 | | Descriptor: | CALCIUM ION, Lipase, ZINC ION | | Authors: | Kanteev, M, Dror, A, Gihaz, S, Fishman, A. | | Deposit date: | 2014-12-09 | | Release date: | 2015-06-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.201 Å) | | Cite: | Structural insights into methanol-stable variants of lipase T6 from Geobacillus stearothermophilus.
Appl.Microbiol.Biotechnol., 99, 2015
|
|
4X85
 
 | | Crystal Structure of lipase from Geobacillus stearothermophilus T6 methanol stable variant H86Y/A269T/R374W | | Descriptor: | CALCIUM ION, Lipase, ZINC ION | | Authors: | Kanteev, M, Dror, A, Gihaz, S, Fishman, A. | | Deposit date: | 2014-12-10 | | Release date: | 2015-06-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.192 Å) | | Cite: | Structural insights into methanol-stable variants of lipase T6 from Geobacillus stearothermophilus.
Appl.Microbiol.Biotechnol., 99, 2015
|
|
6S3G
 
 | | Crystal Structure of lipase from Geobacillus stearothermophilus T6 variant A187C/F291C | | Descriptor: | CALCIUM ION, Lipase, ZINC ION | | Authors: | Gihaz, S, Bash, Y, Rush, I, Shahar, A, Pazy, Y, Fishman, A. | | Deposit date: | 2019-06-25 | | Release date: | 2019-10-09 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Bridges to Stability: Engineering Disulfide Bonds Towards Enhanced Lipase Biodiesel Synthesis
Chemcatchem, 2019
|
|
4Z2R
 
 | |
3NTM
 
 | | Crystal Structure of Tyrosinase from Bacillus megaterium crystallized in the absence of zinc, partial occupancy of CuB | | Descriptor: | COPPER (II) ION, Tyrosinase | | Authors: | Sendovski, M, Kanteev, M, Adir, N, Fishman, A. | | Deposit date: | 2010-07-05 | | Release date: | 2010-11-17 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | First structures of an active bacterial tyrosinase reveal copper plasticity
J.Mol.Biol., 405, 2011
|
|
4Z2T
 
 | |
6FZC
 
 | | Crystal Structure of lipase from Geobacillus stearothermophilus T6 variant L184F/L360F | | Descriptor: | CALCIUM ION, Lipase, ZINC ION | | Authors: | Gihaz, S, Kanteev, M, Pazy, Y, Fishman, A. | | Deposit date: | 2018-03-14 | | Release date: | 2018-10-17 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Filling the Void: Introducing Aromatic Interactions into Solvent Tunnels To Enhance Lipase Stability in Methanol.
Appl.Environ.Microbiol., 84, 2018
|
|
6FZA
 
 | | Crystal Structure of lipase from Geobacillus stearothermophilus T6 methanol stable variant A187F | | Descriptor: | CALCIUM ION, Lipase, ZINC ION | | Authors: | Gihaz, S, Kanteev, M, Pazy, Y, Fishman, A. | | Deposit date: | 2018-03-14 | | Release date: | 2018-10-17 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Filling the Void: Introducing Aromatic Interactions into Solvent Tunnels To Enhance Lipase Stability in Methanol.
Appl.Environ.Microbiol., 84, 2018
|
|
6FZ8
 
 | | Crystal Structure of lipase from Geobacillus stearothermophilus T6 methanol stable variant L184F/A187F | | Descriptor: | CALCIUM ION, Lipase, ZINC ION | | Authors: | Gihaz, S, Kanteev, M, Pazy, Y, Fishman, A. | | Deposit date: | 2018-03-14 | | Release date: | 2018-10-17 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Filling the Void: Introducing Aromatic Interactions into Solvent Tunnels To Enhance Lipase Stability in Methanol.
Appl.Environ.Microbiol., 84, 2018
|
|
6FZD
 
 | | Crystal Structure of lipase from Geobacillus stearothermophilus T6 variant L184F/A187F/L360F | | Descriptor: | CALCIUM ION, Lipase, ZINC ION | | Authors: | Gihaz, S, Kanteev, M, Pazy, Y, Fishman, A. | | Deposit date: | 2018-03-14 | | Release date: | 2018-10-17 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Filling the Void: Introducing Aromatic Interactions into Solvent Tunnels To Enhance Lipase Stability in Methanol.
Appl.Environ.Microbiol., 84, 2018
|
|
