3QTP
 
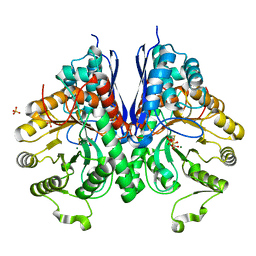 | |
1F3E
 
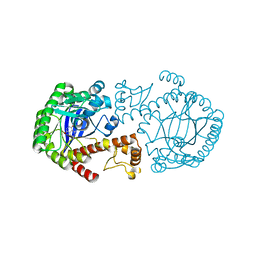 | | A NEW TARGET FOR SHIGELLOSIS: RATIONAL DESIGN AND CRYSTALLOGRAPHIC STUDIES OF INHIBITORS OF TRNA-GUANINE TRANSGLYCOSYLASE | | Descriptor: | 3,5-DIAMINOPHTHALHYDRAZIDE, QUEUINE TRNA-RIBOSYLTRANSFERASE, ZINC ION | | Authors: | Graedler, U, Gerber, H.-D, Goodenough-Lashua, D.M, Garcia, G.A.G, Ficner, R, Reuter, K, Stubbs, M.T, Klebe, G. | | Deposit date: | 2000-06-02 | | Release date: | 2000-06-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | A new target for shigellosis: rational design and crystallographic studies of inhibitors of tRNA-guanine transglycosylase.
J.Mol.Biol., 306, 2001
|
|
6FA9
 
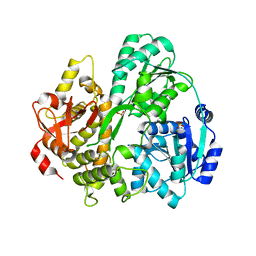 | | CRYSTAL STRUCTURE OF THE DEAH-BOX HELICASE PRP2 | | Descriptor: | Putative mRNA splicing factor, SULFATE ION | | Authors: | Schmitt, A, Hamann, F, Neumann, P, Ficner, R. | | Deposit date: | 2017-12-15 | | Release date: | 2018-07-11 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of the spliceosomal DEAH-box ATPase Prp2.
Acta Crystallogr D Struct Biol, 74, 2018
|
|
6FDF
 
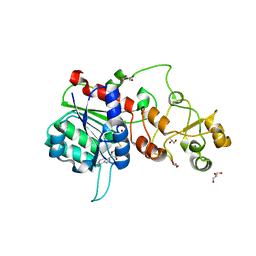 | |
1FJH
 
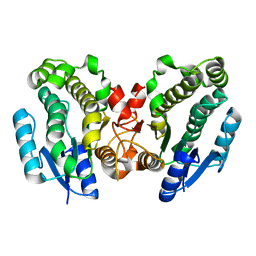 | | THE CRYSTAL STRUCTURE OF 3-ALPHA-HYDROXYSTEROID DEHYDROGENASE FROM COMAMONAS TESTOSTERONI, A MEMBER OF THE SHORT CHAIN DEHYDROGENASE/REDUCTASE FAMILY | | Descriptor: | 3ALPHA-HYDROXYSTEROID DEHYDROGENASE/CARBONYL REDUCTASE | | Authors: | Grimm, C, Ficner, R, Maser, E, Klebe, G, Reuter, K. | | Deposit date: | 2000-08-08 | | Release date: | 2001-01-17 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | The crystal structure of 3alpha -hydroxysteroid dehydrogenase/carbonyl reductase from Comamonas testosteroni shows a novel oligomerization pattern within the short chain dehydrogenase/reductase family.
J.Biol.Chem., 275, 2000
|
|
5LTJ
 
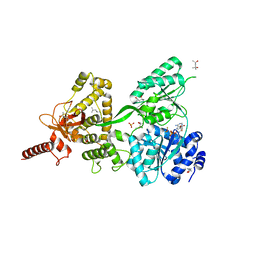 | | Crystal structure of the Prp43-ADP-BeF3 complex (in orthorhombic space group) | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Tauchert, M.J, Ficner, R. | | Deposit date: | 2016-09-06 | | Release date: | 2017-01-25 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Structural insights into the mechanism of the DEAH-box RNA helicase Prp43.
Elife, 6, 2017
|
|
1FK8
 
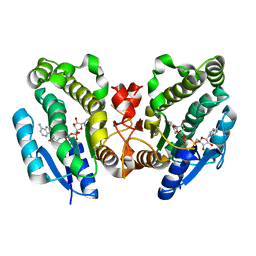 | | THE CRYSTAL STRUCTURE OF THE BINARY COMPLEX WITH NAD OF 3-ALPHA-HYDROXYSTEROID DEHYDROGENASE FROM COMAMONAS TESTOSTERONI, A MEMBER OF THE SHORT CHAIN DEHYDROGENASE/REDUCTASE FAMILY | | Descriptor: | 3ALPHA-HYDROXYSTEROID DEHYDROGENASE/CARBONYL REDUCTASE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Grimm, C, Ficner, R, Maser, E, Klebe, G, Reuter, K. | | Deposit date: | 2000-08-09 | | Release date: | 2001-01-17 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The crystal structure of 3alpha -hydroxysteroid dehydrogenase/carbonyl reductase from Comamonas testosteroni shows a novel oligomerization pattern within the short chain dehydrogenase/reductase family.
J.Biol.Chem., 275, 2000
|
|
5LTA
 
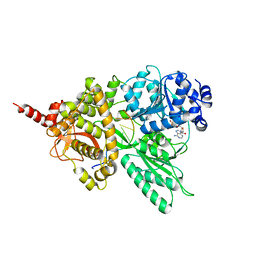 | |
5LTK
 
 | |
6FAC
 
 | | CRYSTAL STRUCTURE OF THE DEAH-BOX HELICASE PRP2 IN COMPLEX WITH ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Schmitt, A, Hamann, F, Neumann, P, Ficner, R. | | Deposit date: | 2017-12-15 | | Release date: | 2018-07-11 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal structure of the spliceosomal DEAH-box ATPase Prp2.
Acta Crystallogr D Struct Biol, 74, 2018
|
|
6FAA
 
 | | CRYSTAL STRUCTURE OF THE DEAH-BOX HELICASE PRP2 IN COMPLEX WITH ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ETHANOL, GLYCEROL, ... | | Authors: | Schmitt, A, Hamann, F, Neumann, P, Ficner, R. | | Deposit date: | 2017-12-15 | | Release date: | 2018-07-11 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Crystal structure of the spliceosomal DEAH-box ATPase Prp2.
Acta Crystallogr D Struct Biol, 74, 2018
|
|
6FA5
 
 | | CRYSTAL STRUCTURE OF THE DEAH-BOX HELICASE PRP2 IN COMPLEX WITH ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DI(HYDROXYETHYL)ETHER, MAGNESIUM ION, ... | | Authors: | Hamann, F, Schmitt, A, Neumann, P, Ficner, R. | | Deposit date: | 2017-12-15 | | Release date: | 2018-07-11 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.303 Å) | | Cite: | Crystal structure of the spliceosomal DEAH-box ATPase Prp2.
Acta Crystallogr D Struct Biol, 74, 2018
|
|
6H42
 
 | | crystal structure of the human TGT catalytic subunit QTRT1 | | Descriptor: | BROMIDE ION, CHLORIDE ION, GLUTAMIC ACID, ... | | Authors: | Johannsson, S, Neumann, P, Ficner, R. | | Deposit date: | 2018-07-20 | | Release date: | 2018-09-05 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Crystal Structure of the Human tRNA Guanine Transglycosylase Catalytic Subunit QTRT1.
Biomolecules, 8, 2018
|
|
3QV2
 
 | |
1S2N
 
 | |
3JQY
 
 | | Crystal Structure of the polySia specific acetyltransferase NeuO | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, Polysialic acid O-acetyltransferase | | Authors: | Schulz, E.-C, Bergfeld, A, Muehlenhoff, M, Ficner, R. | | Deposit date: | 2009-09-08 | | Release date: | 2010-08-18 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.699 Å) | | Cite: | Crystal structure analysis of the polysialic acid specific O-acetyltransferase NeuO
PLoS ONE, 6, 2011
|
|
3I4U
 
 | |
1SH7
 
 | |
4IA6
 
 | | Hydratase from lactobacillus acidophilus in a ligand bound form (LA LAH) | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ... | | Authors: | Khoshnevis, S, Neumann, P, Ficner, R. | | Deposit date: | 2012-12-06 | | Release date: | 2013-03-27 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure analysis of a fatty acid double-bond hydratase from Lactobacillus acidophilus
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4H6B
 
 | | Structural basis for allene oxide cyclization in moss | | Descriptor: | (9Z)-11-{(2R,3S)-3-[(2Z)-pent-2-en-1-yl]oxiran-2-yl}undec-9-enoic acid, (9Z)-11-{(2S,3S)-3-[(2Z)-pent-2-en-1-yl]oxiran-2-yl}undec-9-enoic acid, Allene oxide cyclase, ... | | Authors: | Neumann, P, Ficner, R. | | Deposit date: | 2012-09-19 | | Release date: | 2012-10-17 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Crystal Structures of Physcomitrella patens AOC1 and AOC2: Insights into the Enzyme Mechanism and Differences in Substrate Specificity.
Plant Physiol., 160, 2012
|
|
4H6A
 
 | | Crystal Structure of the Allene Oxide Cyclase 2 from Physcomitrella patens | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, Allene oxide cyclase, ... | | Authors: | Neumann, P, Ficner, R. | | Deposit date: | 2012-09-19 | | Release date: | 2012-10-17 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal Structures of Physcomitrella patens AOC1 and AOC2: Insights into the Enzyme Mechanism and Differences in Substrate Specificity.
Plant Physiol., 160, 2012
|
|
4H6C
 
 | |
4HZK
 
 | |
4IA5
 
 | | Hydratase from Lactobacillus acidophilus - SeMet derivative (apo LAH) | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, GLYCEROL, ... | | Authors: | Khoshnevis, S, Neumann, P, Ficner, R. | | Deposit date: | 2012-12-06 | | Release date: | 2013-03-27 | | Last modified: | 2013-08-07 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Crystal structure analysis of a fatty acid double-bond hydratase from Lactobacillus acidophilus
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4JQ9
 
 | | Dihydrolipoyl dehydrogenase of Escherichia coli pyruvate dehydrogenase complex | | Descriptor: | CHLORIDE ION, Dihydrolipoyl dehydrogenase, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Tietzel, M, Neumann, P, Meyer, D, Ficner, R, Tittmann, K. | | Deposit date: | 2013-03-20 | | Release date: | 2014-04-02 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Dihydrolipoyl dehydrogenase of Escherichia coli pyruvate dehydrogenase complex
TO BE PUBLISHED
|
|
