2BML
 
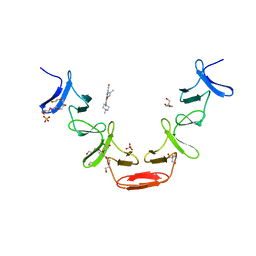 | | Ofloxacin-like antibiotics inhibit pneumococcal cell wall degrading virulence factors | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, AUTOLYSIN, DEXTROFLOXACINE, ... | | Authors: | Fernandez-Tornero, C, Gimenez-Gallego, G, Romero, A, Garcia, E, Pascual-Teresa, B.D, Lopez, R. | | Deposit date: | 2005-03-15 | | Release date: | 2005-03-30 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Ofloxacin-Like Antibiotics Inhibit Pneumococcal Cell Wall-Degrading Virulence Factors
J.Biol.Chem., 280, 2005
|
|
6YD1
 
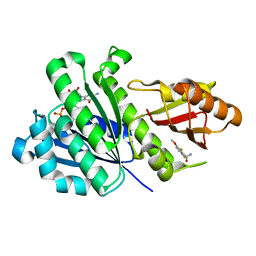 | | SaFtsZ-DFMBA | | Descriptor: | 2,6-difluoro-3-methoxybenzamide, Cell division protein FtsZ, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Fernandez-Tornero, C, Ruiz, F.M, Andreu, J.M. | | Deposit date: | 2020-03-20 | | Release date: | 2021-07-28 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Targeting the FtsZ Allosteric Binding Site with a Novel Fluorescence Polarization Screen, Cytological and Structural Approaches for Antibacterial Discovery.
J.Med.Chem., 64, 2021
|
|
6YD6
 
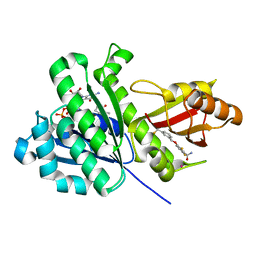 | | SaFtsZ-UCM152 (comp.20) | | Descriptor: | 1,2-ETHANEDIOL, 1-methylpyrrolidin-2-one, 2,6-bis(fluoranyl)-3-[[3-(trifluoromethyl)phenyl]methoxy]benzamide, ... | | Authors: | Fernandez-Tornero, C, Ruiz, F.M, Andreu, J.M. | | Deposit date: | 2020-03-20 | | Release date: | 2021-07-28 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Targeting the FtsZ Allosteric Binding Site with a Novel Fluorescence Polarization Screen, Cytological and Structural Approaches for Antibacterial Discovery.
J.Med.Chem., 64, 2021
|
|
6YD5
 
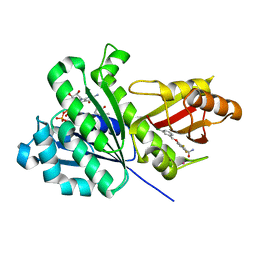 | | SaFtsZ-UCM151 (comp. 18) | | Descriptor: | 1,2-ETHANEDIOL, 1-methylpyrrolidin-2-one, 3-[(3-chlorophenyl)methoxy]-2,6-bis(fluoranyl)benzamide, ... | | Authors: | Fernandez-Tornero, C, Ruiz, F.M, Andreu, J.M. | | Deposit date: | 2020-03-20 | | Release date: | 2021-07-28 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Targeting the FtsZ Allosteric Binding Site with a Novel Fluorescence Polarization Screen, Cytological and Structural Approaches for Antibacterial Discovery.
J.Med.Chem., 64, 2021
|
|
1GVM
 
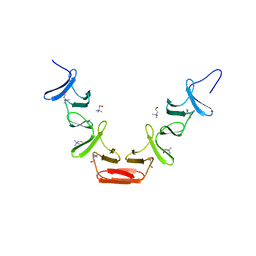 | | CHOLINE BINDING DOMAIN OF THE MAJOR AUTOLYSIN (C-LYTA) FROM STREPTOCOCCUS PNEUMONIAE | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, AUTOLYSIN, CHOLINE ION, ... | | Authors: | Fernandez-Tornero, C, Lopez, R, Garcia, E, Gimenez-Gallego, G, Romero, A. | | Deposit date: | 2002-02-15 | | Release date: | 2002-08-01 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Two New Crystal Forms of the Choline-Binding Domain of the Major Pneumococcal Autolysin: Insights Into the Dynamics of the Active Dimeric
J.Mol.Biol., 321, 2002
|
|
2BYM
 
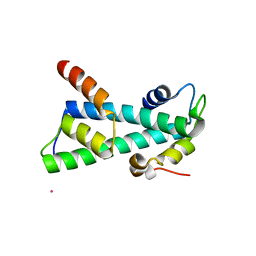 | | Histone fold heterodimer of the Chromatin Accessibility Complex | | Descriptor: | CADMIUM ION, CHRAC-14, CHRAC-16 | | Authors: | Fernandez-Tornero, C, Hartlepp, K.F, Grune, T, Eberharter, A, Becker, P.B, Muller, C.W. | | Deposit date: | 2005-08-03 | | Release date: | 2005-11-09 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The Histone Fold Subunits of Drosophila Chrac Facilitate Nucleosome Sliding Through Dynamic DNA Interactions.
Mol.Cell.Biol., 25, 2005
|
|
2BYK
 
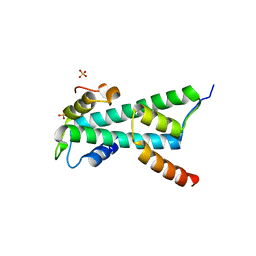 | | Histone fold heterodimer of the Chromatin Accessibility Complex | | Descriptor: | CHRAC-14, CHRAC-16, SULFATE ION | | Authors: | Fernandez-Tornero, C, Hartlepp, K.F, Grune, T, Eberharter, A, Becker, P.B, Muller, C.W. | | Deposit date: | 2005-08-03 | | Release date: | 2005-11-09 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The Histone Fold Subunits of Drosophila Chrac Facilitate Nucleosome Sliding Through Dynamic DNA Interactions.
Mol.Cell.Biol., 25, 2005
|
|
4C3I
 
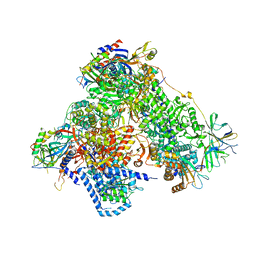 | | Structure of 14-subunit RNA polymerase I at 3.0 A resolution, crystal form C2-100 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, DNA-DIRECTED RNA POLYMERASE I SUBUNIT RPA12, DNA-DIRECTED RNA POLYMERASE I SUBUNIT RPA135, ... | | Authors: | Fernandez-Tornero, C, Moreno-Morcillo, M, Rashid, U.J, Taylor, N.M.I, Ruiz, F.M, Gruene, T, Legrand, P, Steuerwald, U, Muller, C.W. | | Deposit date: | 2013-08-24 | | Release date: | 2013-10-23 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal Structure of the 14-Subunit RNA Polymerase I
Nature, 502, 2013
|
|
4C3H
 
 | | Structure of 14-subunit RNA polymerase I at 3.27 A resolution, crystal form C2-93 | | Descriptor: | DNA-DIRECTED RNA POLYMERASE I SUBUNIT RPA12, DNA-DIRECTED RNA POLYMERASE I SUBUNIT RPA135, DNA-DIRECTED RNA POLYMERASE I SUBUNIT RPA14, ... | | Authors: | Fernandez-Tornero, C, Moreno-Morcillo, M, Rashid, U.J, Taylor, N.M.I, Ruiz, F.M, Gruene, T, Legrand, P, Steuerwald, U, Muller, C.W. | | Deposit date: | 2013-08-24 | | Release date: | 2013-10-23 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.27 Å) | | Cite: | Crystal Structure of the 14-Subunit RNA Polymerase I
Nature, 502, 2013
|
|
4C3J
 
 | | Structure of 14-subunit RNA polymerase I at 3.35 A resolution, crystal form C2-90 | | Descriptor: | DNA-DIRECTED RNA POLYMERASE I SUBUNIT RPA12, DNA-DIRECTED RNA POLYMERASE I SUBUNIT RPA135, DNA-DIRECTED RNA POLYMERASE I SUBUNIT RPA14, ... | | Authors: | Fernandez-Tornero, C, Moreno-Morcillo, M, Rashid, U.J, Taylor, N.M.I, Ruiz, F.M, Gruene, T, Legrand, P, Steuerwald, U, Muller, C.W. | | Deposit date: | 2013-08-24 | | Release date: | 2013-10-23 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.35 Å) | | Cite: | Crystal Structure of the 14-Subunit RNA Polymerase I
Nature, 502, 2013
|
|
1HKN
 
 | | A complex between acidic fibroblast growth factor and 5-amino-2-naphthalenesulfonate | | Descriptor: | 5-AMINO-NAPHTALENE-2-MONOSULFONATE, HEPARIN-BINDING GROWTH FACTOR 1 | | Authors: | Fernandez-Tornero, C, Lozano, R.M, Gimenez-Gallego, G, Romero, A. | | Deposit date: | 2003-03-10 | | Release date: | 2004-03-11 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Leads for Development of New Naphthalenesulfonate Derivatives with Enhanced Antiangiogenic Activity: Crystal Structure of Acidic Fibroblast Growth Factor in Complex with 5-Amino-2-Naphthalenesulfonate
J.Biol.Chem., 278, 2003
|
|
6RVM
 
 | | Cell division protein FtsZ from Staphylococcus aureus, apo form | | Descriptor: | CHLORIDE ION, Cell division protein FtsZ, GLYCEROL, ... | | Authors: | Fernandez-Tornero, C, Andreu, J.M, Canosa-Valls, A.J. | | Deposit date: | 2019-05-31 | | Release date: | 2020-02-19 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.155 Å) | | Cite: | Nucleotide-induced folding of cell division protein FtsZ from Staphylococcus aureus.
Febs J., 287, 2020
|
|
6SI9
 
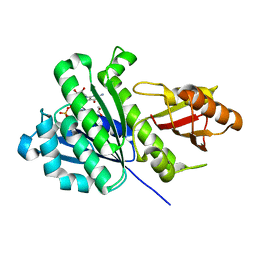 | | FtsZ-refold | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, Cell division protein FtsZ, ... | | Authors: | Fernandez-Tornero, C, Andreu, J.M, Ruiz, F.M. | | Deposit date: | 2019-08-09 | | Release date: | 2020-02-19 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Nucleotide-induced folding of cell division protein FtsZ from Staphylococcus aureus.
Febs J., 287, 2020
|
|
6RVQ
 
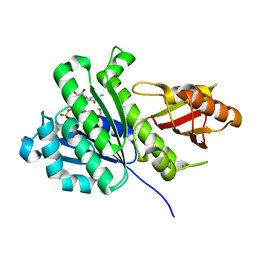 | | SaFtsz-GDP-EthGLy | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, Cell division protein FtsZ, ... | | Authors: | Fernandez-Tornero, C, Andreu, J.M. | | Deposit date: | 2019-05-31 | | Release date: | 2020-02-19 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.136 Å) | | Cite: | Nucleotide-induced folding of cell division protein FtsZ from Staphylococcus aureus.
Febs J., 287, 2020
|
|
6RVN
 
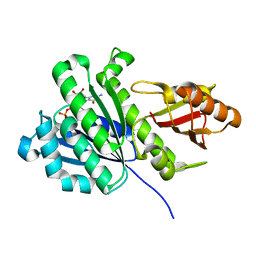 | | aFtsz-GDP-Wat | | Descriptor: | CALCIUM ION, Cell division protein FtsZ, GUANOSINE-5'-DIPHOSPHATE | | Authors: | Fernandez-Tornero, C, Andreu, J.M. | | Deposit date: | 2019-05-31 | | Release date: | 2020-02-19 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.242 Å) | | Cite: | Nucleotide-induced folding of cell division protein FtsZ from Staphylococcus aureus.
Febs J., 287, 2020
|
|
6RVP
 
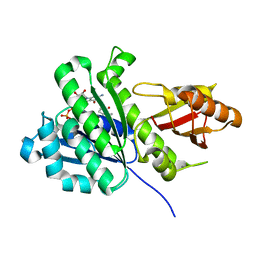 | | SaFtsz-GDP-MetPyr | | Descriptor: | 1-methylpyrrolidin-2-one, CALCIUM ION, Cell division protein FtsZ, ... | | Authors: | Fernandez-Tornero, C, Andreu, J.M. | | Deposit date: | 2019-05-31 | | Release date: | 2020-02-19 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.16 Å) | | Cite: | Nucleotide-induced folding of cell division protein FtsZ from Staphylococcus aureus.
Febs J., 287, 2020
|
|
1HCX
 
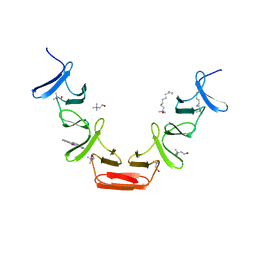 | | Choline binding domain of the major autolysin (C-LytA) from Streptococcus pneumoniae | | Descriptor: | 2,2':6',2''-TERPYRIDINE PLATINUM(II) Chloride, CHOLINE ION, DECYLAMINE-N,N-DIMETHYL-N-OXIDE, ... | | Authors: | Fernandez-Tornero, C, Lopez, R, Garcia, E, Gimenez-Gallego, G, Romero, A. | | Deposit date: | 2001-05-10 | | Release date: | 2001-11-29 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | A Novel Solenoid Fold in the Cell Wall Anchoring Domain of the Pneumococcal Virulence Factor Lyta
Nat.Struct.Biol., 8, 2001
|
|
1H8G
 
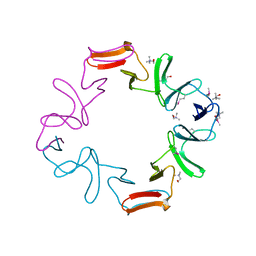 | | C-terminal domain of the major autolysin (C-LytA) from Streptococcus pneumoniae | | Descriptor: | CHOLINE ION, MAJOR AUTOLYSIN | | Authors: | Fernandez-Tornero, C, Garcia, E, Lopez, R, Gimenez-Gallego, G, Romero, A. | | Deposit date: | 2001-02-06 | | Release date: | 2002-01-31 | | Last modified: | 2019-07-24 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | A Novel Solenoid Fold in the Cell Wall Anchoring Domain of the Pneumococcal Virulence Factor Lyta
Nat.Struct.Biol., 8, 2001
|
|
7ON3
 
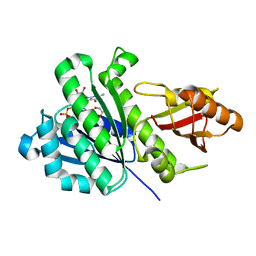 | | SaFtsZ complexed with GDP (soak 10 mM EGTA) | | Descriptor: | 1,2-ETHANEDIOL, Cell division protein FtsZ, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Fernandez-Tornero, C, Ruiz, F.M, Andreu, J.M. | | Deposit date: | 2021-05-25 | | Release date: | 2022-03-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | FtsZ filament structures in different nucleotide states reveal the mechanism of assembly dynamics.
Plos Biol., 20, 2022
|
|
7OJD
 
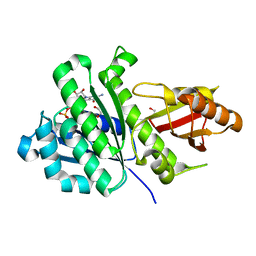 | | SaFtsZ(D46A) complexed with GDP | | Descriptor: | 1,2-ETHANEDIOL, Cell division protein FtsZ, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Fernandez-Tornero, C, Ruiz, F.M, Andreu, J.M. | | Deposit date: | 2021-05-14 | | Release date: | 2022-03-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | FtsZ filament structures in different nucleotide states reveal the mechanism of assembly dynamics.
Plos Biol., 20, 2022
|
|
7OMP
 
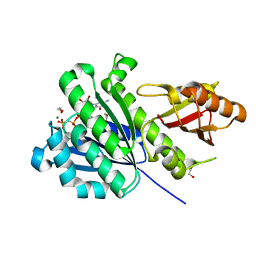 | | SaFtsZ complexed with GDPPCP and Mg2+ | | Descriptor: | 1,2-ETHANEDIOL, Cell division protein FtsZ, MAGNESIUM ION, ... | | Authors: | Fernandez-Tornero, C, Ruiz, F.M, Andreu, J.M. | | Deposit date: | 2021-05-24 | | Release date: | 2022-03-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | FtsZ filament structures in different nucleotide states reveal the mechanism of assembly dynamics.
Plos Biol., 20, 2022
|
|
7OHH
 
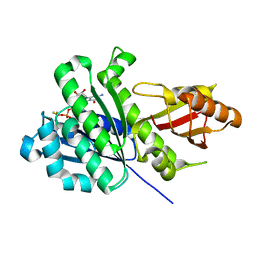 | | SaFtsZ complexed with GDP and BeF3- | | Descriptor: | BERYLLIUM TRIFLUORIDE ION, Cell division protein FtsZ, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Fernandez-Tornero, C, Ruiz, F.M, Andreu, J.M. | | Deposit date: | 2021-05-11 | | Release date: | 2022-03-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | FtsZ filament structures in different nucleotide states reveal the mechanism of assembly dynamics.
Plos Biol., 20, 2022
|
|
7OJA
 
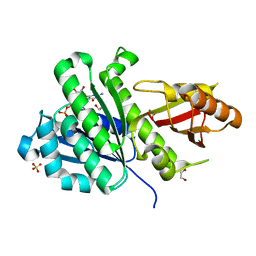 | | SaFtsZ(D210N) complexed with GDP | | Descriptor: | 1,2-ETHANEDIOL, Cell division protein FtsZ, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Fernandez-Tornero, C, Ruiz, F.M, Andreu, J.M. | | Deposit date: | 2021-05-14 | | Release date: | 2022-03-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | FtsZ filament structures in different nucleotide states reveal the mechanism of assembly dynamics.
Plos Biol., 20, 2022
|
|
7ON4
 
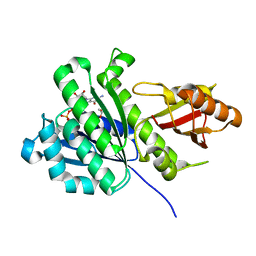 | | SaFtsZ complexed with GDP (co-crystalization with 1mM EDTA) | | Descriptor: | 1,2-ETHANEDIOL, Cell division protein FtsZ, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Fernandez-Tornero, C, Ruiz, F.M, Andreu, J.M. | | Deposit date: | 2021-05-25 | | Release date: | 2022-03-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | FtsZ filament structures in different nucleotide states reveal the mechanism of assembly dynamics.
Plos Biol., 20, 2022
|
|
7OHK
 
 | | SaFtsZ complexed with GDP, BeF3- and Mg2+ | | Descriptor: | BERYLLIUM TRIFLUORIDE ION, Cell division protein FtsZ, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Fernandez-Tornero, C, Ruiz, F.M, Andreu, J.M. | | Deposit date: | 2021-05-11 | | Release date: | 2022-03-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | FtsZ filament structures in different nucleotide states reveal the mechanism of assembly dynamics.
Plos Biol., 20, 2022
|
|
