3S7B
 
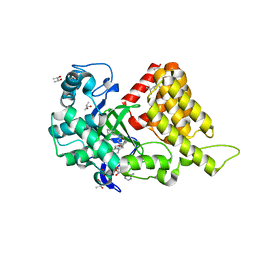 | | Structural Basis of Substrate Methylation and Inhibition of SMYD2 | | Descriptor: | (R,R)-2,3-BUTANEDIOL, N-cyclohexyl-N~3~-[2-(3,4-dichlorophenyl)ethyl]-N-(2-{[2-(5-hydroxy-3-oxo-3,4-dihydro-2H-1,4-benzoxazin-8-yl)ethyl]amino}ethyl)-beta-alaninamide, N-lysine methyltransferase SMYD2, ... | | Authors: | Ferguson, A.D. | | Deposit date: | 2011-05-26 | | Release date: | 2011-08-10 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Structural Basis of Substrate Methylation and Inhibition of SMYD2.
Structure, 19, 2011
|
|
5K76
 
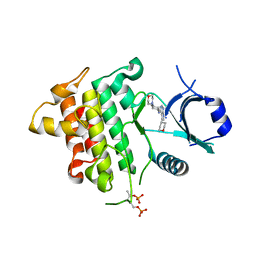 | | IRAK4 in complex with Compound 28 | | Descriptor: | Interleukin-1 receptor-associated kinase 4, ~{N}-(4-morpholin-4-ylcyclohexyl)-5-(oxan-4-yl)-7~{H}-pyrrolo[2,3-d]pyrimidin-4-amine | | Authors: | Ferguson, A.D. | | Deposit date: | 2016-05-25 | | Release date: | 2017-12-06 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.74 Å) | | Cite: | Discovery and Optimization of Pyrrolopyrimidine Inhibitors of Interleukin-1 Receptor Associated Kinase 4 (IRAK4) for the Treatment of Mutant MYD88L265P Diffuse Large B-Cell Lymphoma.
J. Med. Chem., 60, 2017
|
|
5K75
 
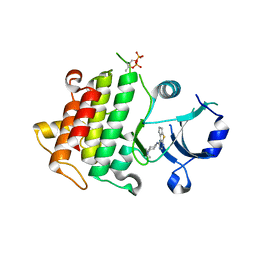 | | IRAK4 in complex with Compound 1 | | Descriptor: | Interleukin-1 receptor-associated kinase 4, SULFATE ION, ~{N}1-(7,8-dihydro-6~{H}-cyclopenta[2,3]thieno[2,4-~{c}]pyrimidin-1-yl)-~{N}4,~{N}4-dimethyl-cyclohexane-1,4-diamine | | Authors: | Ferguson, A.D. | | Deposit date: | 2016-05-25 | | Release date: | 2017-12-06 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Discovery and Optimization of Pyrrolopyrimidine Inhibitors of Interleukin-1 Receptor Associated Kinase 4 (IRAK4) for the Treatment of Mutant MYD88L265P Diffuse Large B-Cell Lymphoma.
J. Med. Chem., 60, 2017
|
|
5KGE
 
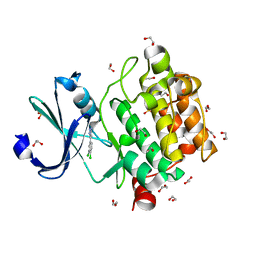 | |
5KGI
 
 | |
5K72
 
 | | IRAK4 in complex with Compound 21 | | Descriptor: | Interleukin-1 receptor-associated kinase 4, SULFATE ION, ~{N}4,~{N}4-dimethyl-~{N}1-[5-(oxan-4-yl)-7~{H}-pyrrolo[2,3-d]pyrimidin-4-yl]cyclohexane-1,4-diamine | | Authors: | Ferguson, A.D. | | Deposit date: | 2016-05-25 | | Release date: | 2017-12-06 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Discovery and Optimization of Pyrrolopyrimidine Inhibitors of Interleukin-1 Receptor Associated Kinase 4 (IRAK4) for the Treatment of Mutant MYD88L265P Diffuse Large B-Cell Lymphoma.
J. Med. Chem., 60, 2017
|
|
5KGG
 
 | |
5K7I
 
 | | IRAK4 in complex with AZ3864 | | Descriptor: | (3~{a}~{R},7~{a}~{S})-1-methyl-5-[4-[[5-(oxan-4-yl)-7~{H}-pyrrolo[2,3-d]pyrimidin-4-yl]amino]cyclohexyl]-3,3~{a},4,6,7,7~{a}-hexahydropyrrolo[3,2-c]pyridin-2-one, Interleukin-1 receptor-associated kinase 4, SULFATE ION | | Authors: | Ferguson, A.D. | | Deposit date: | 2016-05-26 | | Release date: | 2017-12-06 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Discovery and Optimization of Pyrrolopyrimidine Inhibitors of Interleukin-1 Receptor Associated Kinase 4 (IRAK4) for the Treatment of Mutant MYD88L265P Diffuse Large B-Cell Lymphoma.
J. Med. Chem., 60, 2017
|
|
5KGD
 
 | |
5KWH
 
 | | Crystal structure of CK2 | | Descriptor: | 1,2-ETHANEDIOL, Casein kinase II subunit alpha, SULFATE ION, ... | | Authors: | Ferguson, A.D. | | Deposit date: | 2016-07-18 | | Release date: | 2017-10-25 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Crystal structure of CK2
To Be Published
|
|
5KU8
 
 | | Crystal structure of CK2 | | Descriptor: | 1,2-ETHANEDIOL, Casein kinase II subunit alpha, SULFATE ION, ... | | Authors: | Ferguson, A.D, Dowling, J. | | Deposit date: | 2016-07-13 | | Release date: | 2017-11-08 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Crystal structure of CK2
Not Published
|
|
5KU9
 
 | | Crystal structure of MCL1 with compound 1 | | Descriptor: | (3~{S})-3-azanyl-4-(4-bromophenyl)-~{N}-[(3~{S})-1-[2-[[(2~{R})-1-(3,4-dichlorophenyl)-4-(methylamino)-4-oxidanylidene-butan-2-yl]amino]-2-oxidanylidene-ethyl]-2-oxidanylidene-4,5-dihydro-3~{H}-1-benzazepin-3-yl]butanamide, Induced myeloid leukemia cell differentiation protein Mcl-1, SODIUM ION | | Authors: | Ferguson, A.D. | | Deposit date: | 2016-07-13 | | Release date: | 2017-01-11 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure Based Design of Non-Natural Peptidic Macrocyclic Mcl-1 Inhibitors.
ACS Med Chem Lett, 8, 2017
|
|
5K7G
 
 | | IRAK4 in complex with AZ3862 | | Descriptor: | (3~{a}~{S},7~{a}~{R})-1-methyl-5-[4-[[5-(oxan-4-yl)-7~{H}-pyrrolo[2,3-d]pyrimidin-4-yl]amino]cyclohexyl]-3,3~{a},4,6,7,7~{a}-hexahydropyrrolo[3,2-c]pyridin-2-one, Interleukin-1 receptor-associated kinase 4, SULFATE ION | | Authors: | Ferguson, A.D. | | Deposit date: | 2016-05-26 | | Release date: | 2017-12-06 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Discovery and Optimization of Pyrrolopyrimidine Inhibitors of Interleukin-1 Receptor Associated Kinase 4 (IRAK4) for the Treatment of Mutant MYD88L265P Diffuse Large B-Cell Lymphoma.
J. Med. Chem., 60, 2017
|
|
1H4P
 
 | | Crystal structure of exo-1,3-beta glucanse from Saccharomyces cerevisiae | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GLUCAN 1,3-BETA-GLUCOSIDASE I/II, GLYCEROL, ... | | Authors: | Ferguson, A.D. | | Deposit date: | 2001-05-11 | | Release date: | 2003-10-02 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | The Er Protein Folding Sensor Udp-Glucose Glycoprotein:Glucosyltransferase Modifies Substrates Distant to Local Changes in Glycoprotein Conformation.
Nat.Struct.Mol.Biol., 11, 2004
|
|
5J88
 
 | | Structure of the E coli 70S ribosome with the U1060A mutation in 16S rRNA | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,2-ETHANEDIOL, 1,4-DIAMINOBUTANE, ... | | Authors: | Cocozaki, A, Ferguson, A. | | Deposit date: | 2016-04-07 | | Release date: | 2016-07-06 | | Last modified: | 2016-12-07 | | Method: | X-RAY DIFFRACTION (3.32 Å) | | Cite: | Resistance mutations generate divergent antibiotic susceptibility profiles against translation inhibitors.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
7NWK
 
 | | Crystal structure of CDK9-Cyclin T1 bound by compound 6 | | Descriptor: | Cyclin-T1, Cyclin-dependent kinase 9, N-((1R,3R)-3-(7-(4-fluoro-2-methoxyphenyl)-3H-imidazo[4,5-b]pyridin-2-yl)cyclopentyl)acetamide | | Authors: | Collie, G.W, Ferguson, A.D. | | Deposit date: | 2021-03-16 | | Release date: | 2021-10-27 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | Discovery of a Series of 7-Azaindoles as Potent and Highly Selective CDK9 Inhibitors for Transient Target Engagement.
J.Med.Chem., 64, 2021
|
|
4YFN
 
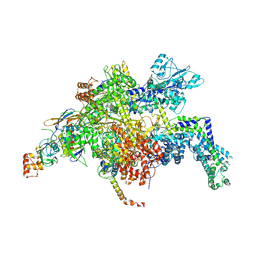 | | Escherichia coli RNA polymerase in complex with squaramide compound 14 (N-[3,4-dioxo-2-(4-{[4-(trifluoromethyl)benzyl]amino}piperidin-1-yl)cyclobut-1-en-1-yl]-3,5-dimethyl-1,2-oxazole-4-sulfonamide) | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Molodtsov, V, Fleming, P.R, Eyermann, C.J, Ferguson, A.D, Foulk, M.A, McKinney, D.C, Masse, C.E, Buurman, E.T, Murakami, K.S. | | Deposit date: | 2015-02-25 | | Release date: | 2015-03-11 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.817 Å) | | Cite: | X-ray Crystal Structures of Escherichia coli RNA Polymerase with Switch Region Binding Inhibitors Enable Rational Design of Squaramides with an Improved Fraction Unbound to Human Plasma Protein.
J.Med.Chem., 58, 2015
|
|
4YFX
 
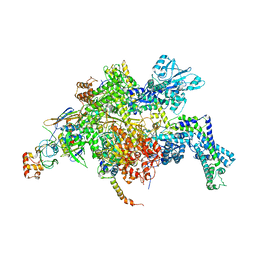 | | Escherichia coli RNA polymerase in complex with Myxopyronin B | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Molodtsov, V, Fleming, P.R, Eyermann, C.J, Ferguson, A.D, Foulk, M.A, McKinney, D.C, Masse, C.E, Buurman, E.T, Murakami, K.S. | | Deposit date: | 2015-02-25 | | Release date: | 2015-03-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.844 Å) | | Cite: | X-ray Crystal Structures of Escherichia coli RNA Polymerase with Switch Region Binding Inhibitors Enable Rational Design of Squaramides with an Improved Fraction Unbound to Human Plasma Protein.
J.Med.Chem., 58, 2015
|
|
4YFK
 
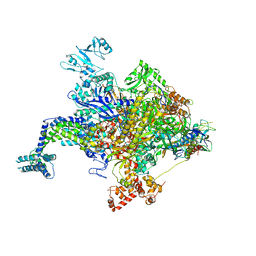 | | Escherichia coli RNA polymerase in complex with squaramide compound 8. | | Descriptor: | 3,5-dimethyl-N-{2-[4-(4-methylbenzyl)piperidin-1-yl]-3,4-dioxocyclobut-1-en-1-yl}-1,2-oxazole-4-sulfonamide, DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Molodtsov, V, Fleming, P.R, Eyermann, C.J, Ferguson, A.D, Foulk, M.A, McKinney, D.C, Masse, C.E, Buurman, E.T, Murakami, K.S. | | Deposit date: | 2015-02-25 | | Release date: | 2015-03-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.571 Å) | | Cite: | X-ray Crystal Structures of Escherichia coli RNA Polymerase with Switch Region Binding Inhibitors Enable Rational Design of Squaramides with an Improved Fraction Unbound to Human Plasma Protein.
J.Med.Chem., 58, 2015
|
|
5N1Z
 
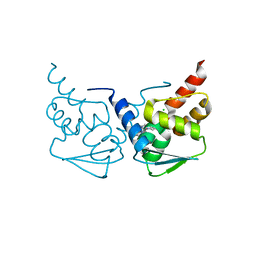 | | Crystal structure of the BCL6 BTB domain in complex with pyrazolo-pyrimidine macrocyclic ligand | | Descriptor: | B-cell lymphoma 6 protein, CHLORIDE ION, pyrazolo-pyrimidine macrocycle | | Authors: | Robb, G, Ferguson, A, Hargreaves, D. | | Deposit date: | 2017-02-06 | | Release date: | 2017-05-17 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Discovery of Pyrazolo[1,5-a]pyrimidine B-Cell Lymphoma 6 (BCL6) Binders and Optimization to High Affinity Macrocyclic Inhibitors.
J. Med. Chem., 60, 2017
|
|
5N1X
 
 | | Crystal structure of the BCL6 BTB domain in complex with pyrazolo-pyrimidine ligand | | Descriptor: | B-cell lymphoma 6 protein, ~{N}-ethyl-5-pyridin-3-yl-pyrazolo[1,5-a]pyrimidin-7-amine | | Authors: | Robb, G, Ferguson, A, Hargreaves, D. | | Deposit date: | 2017-02-06 | | Release date: | 2017-05-17 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Discovery of Pyrazolo[1,5-a]pyrimidine B-Cell Lymphoma 6 (BCL6) Binders and Optimization to High Affinity Macrocyclic Inhibitors.
J. Med. Chem., 60, 2017
|
|
5N20
 
 | | Crystal structure of the BCL6 BTB domain in complex with pyrazolo-pyrimidine ligand | | Descriptor: | B-cell lymphoma 6 protein, ~{N}-[5-[[3-cyano-7-(cyclopropylamino)pyrazolo[1,5-a]pyrimidin-5-yl]amino]-2-[(3~{R})-3-(dimethylamino)pyrrolidin-1-yl]phenyl]ethanamide | | Authors: | Robb, G, Ferguson, A, Hargreaves, D. | | Deposit date: | 2017-02-06 | | Release date: | 2017-05-17 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | Discovery of Pyrazolo[1,5-a]pyrimidine B-Cell Lymphoma 6 (BCL6) Binders and Optimization to High Affinity Macrocyclic Inhibitors.
J. Med. Chem., 60, 2017
|
|
6EW6
 
 | | Crystal structure of the BCL6 BTB domain in complex with anilinopyrimidine ligand | | Descriptor: | B-cell lymphoma 6 protein, ~{N}2-(2-chlorophenyl)-1,3,5-triazine-2,4-diamine | | Authors: | Robb, G, Ferguson, A, Hargreaves, D. | | Deposit date: | 2017-11-03 | | Release date: | 2018-10-24 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | Development of a Novel B-Cell Lymphoma 6 (BCL6) PROTAC To Provide Insight into Small Molecule Targeting of BCL6.
ACS Chem. Biol., 13, 2018
|
|
6EW8
 
 | |
6EW7
 
 | |
