7YVB
 
 | | Aplysia californica FaNaC in ligand bound state | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, FMRFamide-gated Na+ channel, ... | | Authors: | Chen, Q.F, Liu, F.L, Dang, Y, Feng, H, Zhang, Z, Ye, S. | | Deposit date: | 2022-08-19 | | Release date: | 2023-08-09 | | Last modified: | 2023-10-11 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structure and mechanism of a neuropeptide-activated channel in the ENaC/DEG superfamily.
Nat.Chem.Biol., 19, 2023
|
|
2LY8
 
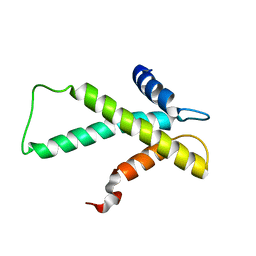 | | The budding yeast chaperone Scm3 recognizes the partially unfolded dimer of the centromere-specific Cse4/H4 histone variant | | Descriptor: | Budding yeast chaperone Scm3 | | Authors: | Hong, J, Feng, H, Zhou, Z, Ghirlando, R, Bai, Y. | | Deposit date: | 2012-09-13 | | Release date: | 2012-12-12 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Identification of Functionally Conserved Regions in the Structure of the Chaperone/CenH3/H4 Complex.
J.Mol.Biol., 425, 2013
|
|
2MZD
 
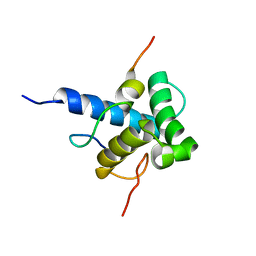 | | Characterization of the p300 Taz2-p53 TAD2 Complex and Comparison with the p300 Taz2-p53 TAD1 Complex | | Descriptor: | Cellular tumor antigen p53, Histone acetyltransferase p300 | | Authors: | Miller Jenkins, L.M, Feng, H, Durell, S.R, Tagad, H.D, Mazur, S.J, Tropea, J.E, Bai, Y, Appella, E. | | Deposit date: | 2015-02-11 | | Release date: | 2015-03-25 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Characterization of the p300 Taz2-p53 TAD2 Complex and Comparison with the p300 Taz2-p53 TAD1 Complex.
Biochemistry, 54, 2015
|
|
2K8F
 
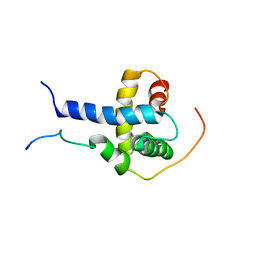 | | Structural Basis for the Regulation of p53 Function by p300 | | Descriptor: | Cellular tumor antigen p53, Histone acetyltransferase p300 | | Authors: | Bai, Y, Feng, H, Jenkins, L.M, Durell, S.R, Wiodawer, A, Appella, E. | | Deposit date: | 2008-09-08 | | Release date: | 2009-03-03 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structural Basis for p300 Taz2-p53 TAD1 Binding and Modulation by Phosphorylation.
Structure, 17, 2009
|
|
5X6D
 
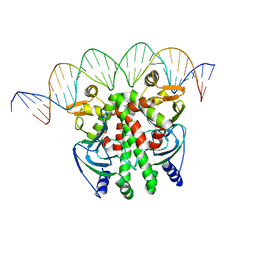 | | Crystal structure of PrfA-DNA binary complex | | Descriptor: | DNA (28-MER), DNA (29-MER), Listeriolysin positive regulatory factor A | | Authors: | Wang, Y, Feng, H, Zhu, Y.L, Gao, P. | | Deposit date: | 2017-02-21 | | Release date: | 2017-04-12 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.94 Å) | | Cite: | Structural insights into glutathione-mediated activation of the master regulator PrfA in Listeria monocytogenes
Protein Cell, 8, 2017
|
|
5X6E
 
 | | Crystal structure of PrfA-DNA binary complex | | Descriptor: | DNA (28-MER), DNA (29-MER), GLUTATHIONE, ... | | Authors: | Wang, Y, Feng, H, Zhu, Y.L, Gao, P. | | Deposit date: | 2017-02-21 | | Release date: | 2017-04-12 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.99 Å) | | Cite: | Structural insights into glutathione-mediated activation of the master regulator PrfA in Listeria monocytogenes
Protein Cell, 8, 2017
|
|
