4RG8
 
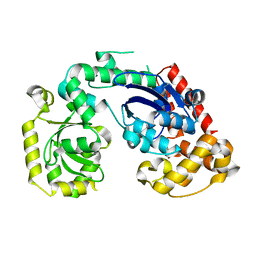 | | Structural and biochemical studies of a moderately thermophilic Exonuclease I from Methylocaldum szegediense | | Descriptor: | Exonuclease I, MAGNESIUM ION | | Authors: | Fei, L, Tian, S, Moysey, R, Misca, M, Barker, J.J, Smith, M.A, McEwan, P.A, Pilka, E.S, Crawley, L, Evans, T, Sun, D. | | Deposit date: | 2014-09-29 | | Release date: | 2015-02-25 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Structural and Biochemical Studies of a Moderately Thermophilic Exonuclease I from Methylocaldum szegediense.
Plos One, 10, 2015
|
|
5OU8
 
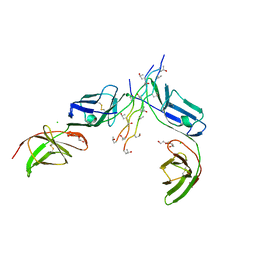 | |
8EVG
 
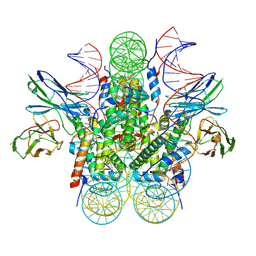 | |
7X3T
 
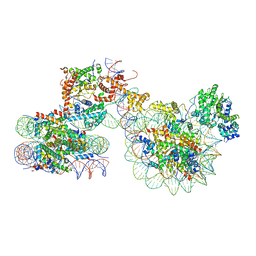 | | Cryo-EM structure of ISW1a-dinucleosome | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, DNA (343-MER), ... | | Authors: | Lifei, L, Kangjing, C, Chen, Z. | | Deposit date: | 2022-03-01 | | Release date: | 2023-09-20 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (5.4 Å) | | Cite: | Structure of the ISW1a complex bound to the dinucleosome.
Nat.Struct.Mol.Biol., 31, 2024
|
|
7X3W
 
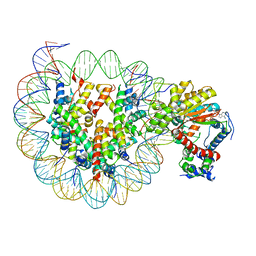 | | Cryo-EM structure of ISW1-N1 nucleosome | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, DNA (146-MER), ... | | Authors: | Lifei, L, Kangjing, C, Chen, Z. | | Deposit date: | 2022-03-01 | | Release date: | 2023-09-20 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structure of the ISW1a complex bound to the dinucleosome.
Nat.Struct.Mol.Biol., 31, 2024
|
|
7X3X
 
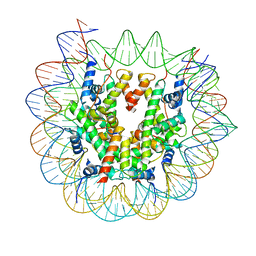 | | Cryo-EM structure of N1 nucleosome-RA | | Descriptor: | DNA (146-MER), Histone H2A, Histone H2B 1.1, ... | | Authors: | Lifei, L, Kangjing, C, Chen, Z. | | Deposit date: | 2022-03-01 | | Release date: | 2023-09-20 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structure of the ISW1a complex bound to the dinucleosome.
Nat.Struct.Mol.Biol., 31, 2024
|
|
7X3V
 
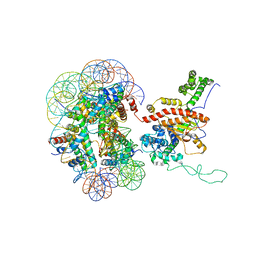 | | Cryo-EM structure of IOC3-N2 nucleosome | | Descriptor: | DNA (146-MER), Histone H2A, Histone H2B 1.1, ... | | Authors: | Lifei, L, Kangjing, C, Chen, Z. | | Deposit date: | 2022-03-01 | | Release date: | 2023-09-20 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (3.09 Å) | | Cite: | Structure of the ISW1a complex bound to the dinucleosome.
Nat.Struct.Mol.Biol., 31, 2024
|
|
5D3D
 
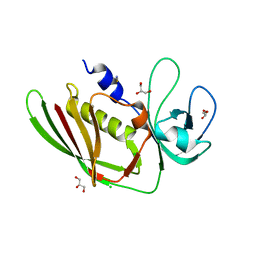 | |
5D3I
 
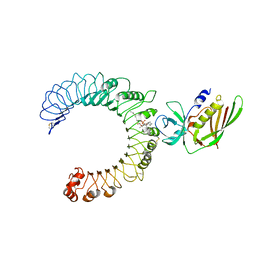 | | Crystal structure of the SSL3-TLR2 complex | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Feitsma, L.J, Huizinga, E.G. | | Deposit date: | 2015-08-06 | | Release date: | 2015-08-19 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural basis for inhibition of TLR2 by staphylococcal superantigen-like protein 3 (SSL3).
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
7U0I
 
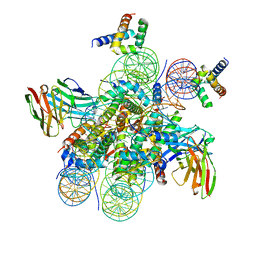 | | Structure of LIN28b nucleosome bound 2 OCT4 | | Descriptor: | DNA (162-MER), Histone H2A type 2-C, Histone H2B type 2-E, ... | | Authors: | Tengfei, L, Guan, R, Bai, Y. | | Deposit date: | 2022-02-18 | | Release date: | 2023-06-28 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Structural mechanism of LIN28B nucleosome targeting by OCT4.
Mol.Cell, 83, 2023
|
|
5NQH
 
 | | Structure of the human Fe65-PTB2 homodimer | | Descriptor: | Amyloid beta A4 precursor protein-binding family B member 1, GLYCEROL, SULFATE ION | | Authors: | Feilen, L.P, Haubrich, K, Sinning, I, Konietzko, U, Kins, S, Simon, B, Wild, K. | | Deposit date: | 2017-04-20 | | Release date: | 2017-05-03 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Fe65-PTB2 Dimerization Mimics Fe65-APP Interaction.
Front Mol Neurosci, 10, 2017
|
|
5OU7
 
 | |
5OU9
 
 | |
2QE4
 
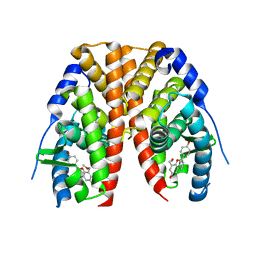 | | Estrogen receptor alpha ligand-binding domain in complex with a benzopyran agonist | | Descriptor: | (3AS,4R,9BR)-4-(4-HYDROXYPHENYL)-6-(METHOXYMETHYL)-1,2,3,3A,4,9B-HEXAHYDROCYCLOPENTA[C]CHROMEN-8-OL, Estrogen receptor | | Authors: | Norman, B.H, Richardson, T.I, Dodge, J.A, Pfeifer, L.A, Durst, G.L, Wang, Y, Durbin, J.D, Krishnan, V, Dinn, S.R, Liu, S.Q, Reilly, J.E, Ryter, K.T. | | Deposit date: | 2007-06-22 | | Release date: | 2007-09-04 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Benzopyrans as selective estrogen receptor beta agonists (SERBAs). Part 4: Functionalization of the benzopyran A-ring.
Bioorg.Med.Chem.Lett., 17, 2007
|
|
5JIF
 
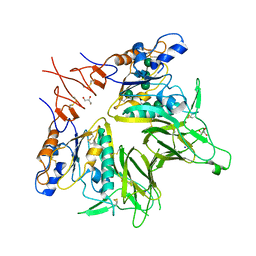 | | Crystal structure of mouse hepatitis virus strain DVIM Hemagglutinin-Esterase | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, ... | | Authors: | Zeng, Q.H, Bakkers, M.J.G, Feitsma, L.J, de Groot, R.J, Huizinga, E.G. | | Deposit date: | 2016-04-22 | | Release date: | 2016-05-11 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Coronavirus receptor switch explained from the stereochemistry of protein-carbohydrate interactions and a single mutation.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5JIL
 
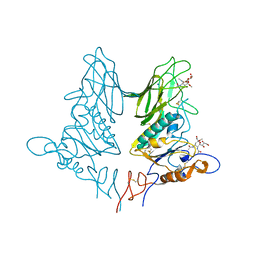 | | Crystal structure of rat coronavirus strain New-Jersey Hemagglutinin-Esterase in complex with 4N-acetyl sialic acid | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin-esterase, ... | | Authors: | Bakkers, M.J.G, Feitsma, L.J, de Groot, R.J, Huizinga, E.G. | | Deposit date: | 2016-04-22 | | Release date: | 2016-05-11 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Coronavirus receptor switch explained from the stereochemistry of protein-carbohydrate interactions and a single mutation.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5N11
 
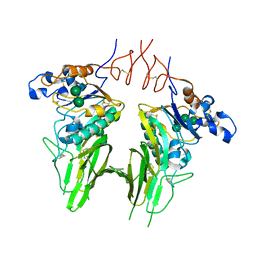 | | Crystal structure of Human beta1-coronavirus OC43 NL/A/2005 Hemagglutinin-Esterase | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ACETIC ACID, ... | | Authors: | Bakkers, M.J.G, Feitsma, L.J, de Groot, R.J, Huizinga, E.G. | | Deposit date: | 2017-02-04 | | Release date: | 2017-03-22 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Betacoronavirus Adaptation to Humans Involved Progressive Loss of Hemagglutinin-Esterase Lectin Activity.
Cell Host Microbe, 21, 2017
|
|
4ZXN
 
 | | Crystal structure of rat coronavirus strain New-Jersey Hemagglutinin-Esterase | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, HE protein, SODIUM ION, ... | | Authors: | Bakkers, M.J.G, Feitsma, L.J, Huizinga, E.G, de Groot, R.J. | | Deposit date: | 2015-05-20 | | Release date: | 2016-05-11 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structures of murine coronavirus hemagglutinin-esterases reveal structural basis for esterase substrate specificity
To Be Published
|
|
2JJ3
 
 | | Estrogen receptor beta ligand binding domain in complex with a Benzopyran agonist | | Descriptor: | (3AS,4R,9BR)-4-(4-HYDROXYPHENYL)-6-(METHOXYMETHYL)-1,2,3,3A,4,9B-HEXAHYDROCYCLOPENTA[C]CHROMEN-8-OL, ESTROGEN RECEPTOR BETA | | Authors: | Norman, B.H, Richardson, T.I, Dodge, J.A, Pfeifer, L.A, Durst, G.L, Wang, Y, Durbin, J.D, Krishnan, V, Dinn, S.R, Liu, S, Reilly, J.E, Ryter, K.T. | | Deposit date: | 2007-07-03 | | Release date: | 2007-08-07 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Benzopyrans as Selective Estrogen Receptor Beta Agonists (Serbas). Part 4: Functionalization of the Benzopyran A-Ring.
Bioorg.Med.Chem.Lett., 17, 2007
|
|
