3CAQ
 
 | | Crystal structure of 5beta-reductase (AKR1D1) in complex with NADPH | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,2-ETHANEDIOL, 3-oxo-5-beta-steroid 4-dehydrogenase, ... | | Authors: | Faucher, F, Cantin, L, Breton, R. | | Deposit date: | 2008-02-20 | | Release date: | 2008-12-09 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structures of Human Delta4-3-Ketosteroid 5beta-Reductase (AKR1D1) Reveal the Presence of an Alternative Binding Site Responsible for Substrate Inhibition (dagger) (,) (double dagger).
Biochemistry, 47, 2008
|
|
3KNT
 
 | |
3I0X
 
 | |
3I0W
 
 | |
4GAC
 
 | |
3CAV
 
 | | Crystal structure of 5beta-reductase (AKR1D1) in complex with NADP+ and 5beta-pregnan-3,20-dione | | Descriptor: | (5BETA)-PREGNANE-3,20-DIONE, 1,2-ETHANEDIOL, 3-oxo-5-beta-steroid 4-dehydrogenase, ... | | Authors: | Faucher, F, Cantin, L, Breton, R. | | Deposit date: | 2008-02-20 | | Release date: | 2008-07-29 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The crystal structure of human Delta4-3-ketosteroid 5beta-reductase defines the functional role of the residues of the catalytic tetrad in the steroid double bond reduction mechanism.
Biochemistry, 47, 2008
|
|
3CAS
 
 | | Crystal structure of 5beta-reductase (AKR1D1) in complex with NADP+ and 4-androstenedione | | Descriptor: | 1,2-ETHANEDIOL, 3-oxo-5-beta-steroid 4-dehydrogenase, 4-ANDROSTENE-3-17-DIONE, ... | | Authors: | Faucher, F, Cantin, L, Breton, R. | | Deposit date: | 2008-02-20 | | Release date: | 2008-12-09 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structures of Human Delta4-3-Ketosteroid 5beta-Reductase (AKR1D1) Reveal the Presence of an Alternative Binding Site Responsible for Substrate Inhibition (dagger) (,) (double dagger).
Biochemistry, 47, 2008
|
|
2HE8
 
 | | Crystal structure of 17alpha-hydroxysteroid dehydrogenase in its apo-form | | Descriptor: | ACETATE ION, Aldo-keto reductase family 1, member C21, ... | | Authors: | Faucher, F, Pereira de Jesus-Tran, K, Cantin, L, Luu-the, V, Labrie, F, Breton, R. | | Deposit date: | 2006-06-21 | | Release date: | 2006-12-05 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structures of Mouse 17alpha-Hydroxysteroid Dehydrogenase (Apoenzyme and Enzyme-NADP(H) Binary Complex): Identification of Molecular Determinants Responsible for the Unique 17alpha-reductive Activity of this Enzyme.
J.Mol.Biol., 364, 2006
|
|
2HDJ
 
 | | Crystal structure of human type 3 3alpha-hydroxysteroid dehydrogenase in complex with NADP(H) | | Descriptor: | 1,2-ETHANEDIOL, Aldo-keto reductase family 1 member C2, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Faucher, F, Pereira de Jesus-Tran, K, Cantin, L, Luu-the, V, Labrie, F, Breton, R. | | Deposit date: | 2006-06-20 | | Release date: | 2006-12-05 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structures of Mouse 17alpha-Hydroxysteroid Dehydrogenase (Apoenzyme and Enzyme-NADP(H) Binary Complex): Identification of Molecular Determinants Responsible for the Unique 17alpha-reductive Activity of this Enzyme.
J.Mol.Biol., 364, 2006
|
|
2HE5
 
 | | Crystal structure of 17alpha-hydroxysteroid dehydrogenase in binary complex with NADP(H) in an open conformation | | Descriptor: | ACETATE ION, Aldo-keto reductase family 1, member C21, ... | | Authors: | Faucher, F, Pereira de Jesus-Tran, K, Cantin, L, Luu-the, V, Labrie, F, Breton, R. | | Deposit date: | 2006-06-21 | | Release date: | 2006-12-05 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal Structures of Mouse 17alpha-Hydroxysteroid Dehydrogenase (Apoenzyme and Enzyme-NADP(H) Binary Complex): Identification of Molecular Determinants Responsible for the Unique 17alpha-reductive Activity of this Enzyme.
J.Mol.Biol., 364, 2006
|
|
2IPG
 
 | | Crystal structure of 17alpha-hydroxysteroid dehydrogenase mutant K31A in complex with NADP+ and epi-testosterone | | Descriptor: | (10ALPHA,13ALPHA,14BETA,17ALPHA)-17-HYDROXYANDROST-4-EN-3-ONE, 1,2-ETHANEDIOL, 3(17)alpha-hydroxysteroid dehydrogenase, ... | | Authors: | Faucher, F, Cantin, L, Pereira de Jesus-Tran, K, Lemieux, M, Luu-the, V, Labrie, F, Breton, R. | | Deposit date: | 2006-10-12 | | Release date: | 2007-06-19 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Mouse 17alpha-Hydroxysteroid Dehydrogenase (AKR1C21) Binds Steroids Differently from other Aldo-keto Reductases: Identification and Characterization of Amino Acid Residues Critical for Substrate Binding.
J.Mol.Biol., 369, 2007
|
|
2IPJ
 
 | | Crystal structure of h3alpha-hydroxysteroid dehydrogenase type 3 mutant Y24A in complex with NADP+ and epi-testosterone | | Descriptor: | (10ALPHA,13ALPHA,14BETA,17ALPHA)-17-HYDROXYANDROST-4-EN-3-ONE, 1,2-ETHANEDIOL, Aldo-keto reductase family 1 member C2, ... | | Authors: | Faucher, F, Cantin, L, Pereira de Jesus-Tran, K, Luu-the, V, Labrie, F, Breton, R. | | Deposit date: | 2006-10-12 | | Release date: | 2007-06-19 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Mouse 17alpha-Hydroxysteroid Dehydrogenase (AKR1C21) Binds Steroids Differently from other Aldo-keto Reductases: Identification and Characterization of Amino Acid Residues Critical for Substrate Binding.
J.Mol.Biol., 369, 2007
|
|
2IPF
 
 | | Crystal structure of 17alpha-hydroxysteroid dehydrogenase in complex with NADP+ and epi-testosterone | | Descriptor: | (10ALPHA,13ALPHA,14BETA,17ALPHA)-17-HYDROXYANDROST-4-EN-3-ONE, (3(17)alpha-hydroxysteroid dehydrogenase), 1,2-ETHANEDIOL, ... | | Authors: | Faucher, F, Cantin, L, Pereira de Jesus-Tran, K, Luu-the, V, Labrie, F, Breton, R. | | Deposit date: | 2006-10-12 | | Release date: | 2007-06-19 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Mouse 17alpha-Hydroxysteroid Dehydrogenase (AKR1C21) Binds Steroids Differently from other Aldo-keto Reductases: Identification and Characterization of Amino Acid Residues Critical for Substrate Binding.
J.Mol.Biol., 369, 2007
|
|
3FHG
 
 | |
2HEJ
 
 | | Crystal structure of 17alpha-hydroxysteroid dehydrogenase in complex with NADP(H) in a closed conformation | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,2-ETHANEDIOL, Aldo-keto reductase family 1, ... | | Authors: | Faucher, F, Pereira de Jesus-Tran, K, Cantin, L, Luu-the, V, Labrie, F, Breton, R. | | Deposit date: | 2006-06-21 | | Release date: | 2006-12-05 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Crystal Structures of Mouse 17alpha-Hydroxysteroid Dehydrogenase (Apoenzyme and Enzyme-NADP(H) Binary Complex): Identification of Molecular Determinants Responsible for the Unique 17alpha-reductive Activity of this Enzyme.
J.Mol.Biol., 364, 2006
|
|
3F0Z
 
 | |
3FHF
 
 | |
3F10
 
 | |
2PNU
 
 | | Crystal structure of human androgen receptor ligand-binding domain in complex with EM-5744 | | Descriptor: | (5S,8R,9S,10S,13R,14S,17S)-13-{2-[(3,5-DIFLUOROBENZYL)OXY]ETHYL}-17-HYDROXY-10-METHYLHEXADECAHYDRO-3H-CYCLOPENTA[A]PHENANTHREN-3-ONE, 2,3-DIHYDROXY-1,4-DITHIOBUTANE, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ... | | Authors: | Cantin, L, Faucher, F, Couture, J.F, Pereira de Jesus-Tran, K, Legrand, P, Ciobanu, C.L, Singh, S.M, Labrie, F, Breton, R. | | Deposit date: | 2007-04-25 | | Release date: | 2007-09-11 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural Characterization of the Human Androgen Receptor Ligand-binding Domain Complexed with EM5744, a Rationally Designed Steroidal Ligand Bearing a Bulky Chain Directed toward Helix 12.
J.Biol.Chem., 282, 2007
|
|
5BW0
 
 | | The crystal structure of minor pseudopilin binary complex of XcpV and XcpW from the Type 2 secretion system of Pseudomonas aeruginosa | | Descriptor: | SULFATE ION, Type II secretion system protein I, Type II secretion system protein J | | Authors: | Zhang, Y, Faucher, F, Poole, K, Jia, Z. | | Deposit date: | 2015-06-05 | | Release date: | 2016-07-20 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure-guided disruption of the pseudopilus tip complex inhibits the Type II secretion in Pseudomonas aeruginosa.
PLoS Pathog., 14, 2018
|
|
3P2U
 
 | | Crystal structure of PhnP in complex with orthovanadate | | Descriptor: | MANGANESE (II) ION, Protein phnP, VANADATE ION, ... | | Authors: | Podzelinska, K, Kim, Y.M, Zechel, D, Faucher, F, Jia, Z. | | Deposit date: | 2010-10-04 | | Release date: | 2011-01-05 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Crystal structure of PhnP in complex with orthovanadate
TO BE PUBLISHED
|
|
3ULT
 
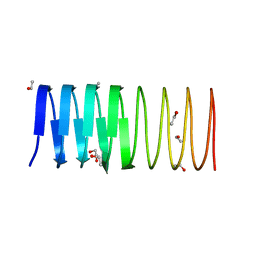 | | Crystal structure of an ice-binding protein from the perennial ryegrass, Lolium perenne | | Descriptor: | 1,2-ETHANEDIOL, ETHANOL, Ice recrystallization inhibition protein-like protein | | Authors: | Middleton, A.J, Faucher, F, Campbell, R.L, Davies, P.L. | | Deposit date: | 2011-11-11 | | Release date: | 2012-02-15 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Antifreeze protein from freeze-tolerant grass has a beta-roll fold with an irregularly structured ice-binding site.
J.Mol.Biol., 416, 2012
|
|
4IL1
 
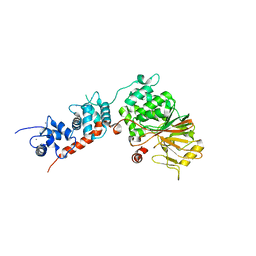 | | Crystal Structure of the Rat Calcineurin | | Descriptor: | CALCIUM ION, Calmodulin, Calcineurin subunit B type 1, ... | | Authors: | Ye, Q, Faucher, F, Jia, Z. | | Deposit date: | 2012-12-28 | | Release date: | 2013-10-02 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural basis of calcineurin activation by calmodulin.
Cell Signal, 25, 2013
|
|
4FL4
 
 | | Scaffoldin conformation and dynamics revealed by a ternary complex from the Clostridium thermocellum cellulosome | | Descriptor: | CALCIUM ION, Cellulosome anchoring protein cohesin region, Glycoside hydrolase family 9, ... | | Authors: | Currie, M.A, Adams, J.J, Faucher, F, Bayer, E.A, Jia, Z, Smith, S.P, Montreal-Kingston Bacterial Structural Genomics Initiative (BSGI) | | Deposit date: | 2012-06-14 | | Release date: | 2012-06-27 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Scaffoldin Conformation and Dynamics Revealed by a Ternary Complex from the Clostridium thermocellum Cellulosome.
J.Biol.Chem., 287, 2012
|
|
