6ADW
 
 | | Crystal structure of the Zika virus NS3 helicase (apo form) | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, SULFATE ION, Serine protease NS3 | | Authors: | Fang, J, Lu, G, Gong, P. | | Deposit date: | 2018-08-02 | | Release date: | 2019-03-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystallographic Snapshots of the Zika Virus NS3 Helicase Help Visualize the Reactant Water Replenishment.
ACS Infect Dis, 5, 2019
|
|
6ADX
 
 | | Crystal structure of the Zika virus NS3 helicase (ADP-Mn2+ complex, form 1) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MANGANESE (II) ION, Serine protease NS3 | | Authors: | Fang, J, Lu, G, Gong, P. | | Deposit date: | 2018-08-02 | | Release date: | 2019-03-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.752 Å) | | Cite: | Crystallographic Snapshots of the Zika Virus NS3 Helicase Help Visualize the Reactant Water Replenishment.
ACS Infect Dis, 5, 2019
|
|
6ADY
 
 | | Crystal structure of the Zika virus NS3 helicase (ADP-Mn2+ complex, form 2) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MANGANESE (II) ION, Serine protease NS3 | | Authors: | Fang, J, Lu, G, Gong, P. | | Deposit date: | 2018-08-02 | | Release date: | 2019-03-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystallographic Snapshots of the Zika Virus NS3 Helicase Help Visualize the Reactant Water Replenishment.
ACS Infect Dis, 5, 2019
|
|
7UBU
 
 | |
3BS8
 
 | | Crystal structure of Glutamate 1-Semialdehyde Aminotransferase complexed with pyridoxamine-5'-phosphate From Bacillus subtilis | | Descriptor: | 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, Glutamate-1-semialdehyde 2,1-aminomutase | | Authors: | Ge, H, Fan, J, Teng, M, Niu, L. | | Deposit date: | 2007-12-22 | | Release date: | 2008-12-23 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of Glutamate1-semialdehyde aminotransferase from Bacillus subtilis with bound pyridoxamine-5'-phosphate
Biochem.Biophys.Res.Commun., 402, 2010
|
|
8K6Z
 
 | | NMR structure of human leptin | | Descriptor: | Leptin | | Authors: | Fan, X, Qin, R, Yuan, W, Fan, J, Huang, W, Lin, Z. | | Deposit date: | 2023-07-26 | | Release date: | 2024-02-07 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The solution structure of human leptin reveals a conformational plasticity important for receptor recognition.
Structure, 32, 2024
|
|
6LCI
 
 | | Solution structure of mdaA-1 domain | | Descriptor: | mdaA-1 | | Authors: | Nguyen, T.A, Le, S, Lee, M, Fan, J.S, Yang, D, Yan, J, Jedd, G. | | Deposit date: | 2019-11-19 | | Release date: | 2020-11-25 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Fungal Wound Healing through Instantaneous Protoplasmic Gelation.
Curr.Biol., 31, 2021
|
|
6KYL
 
 | | Crystal Structure of Phosphatidic acid Transporter Ups1/Mdm35 in Complex with (2R)-3-(phosphonooxy)propane-1,2-diyl dihexanoate | | Descriptor: | (2R)-3-(phosphonooxy)propane-1,2-diyl dihexanoate, Mitochondrial distribution and morphology protein 35, Protein UPS1, ... | | Authors: | Lu, J, Chan, K.C, Zhai, Y, Fan, J, Sun, F. | | Deposit date: | 2019-09-19 | | Release date: | 2019-10-02 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.55 Å) | | Cite: | Molecular mechanism of mitochondrial phosphatidate transfer by Ups1.
Commun Biol, 3, 2020
|
|
4CKH
 
 | | Helical reconstruction of ACAP1(BAR-PH domain) decorated membrane tubules by cryo-electron microscopy | | Descriptor: | ARF-GAP WITH COILED-COIL, ANK REPEAT AND PH DOMAIN-CONTAINING PROTEIN 1 | | Authors: | Pang, X.Y, Fan, J, Zhang, Y, Zhang, K, Gao, B.Q, Ma, J, Li, J, Deng, Y.C, Zhou, Q.J, Hsu, V, Sun, F. | | Deposit date: | 2014-01-06 | | Release date: | 2014-10-15 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (17 Å) | | Cite: | A Ph Domain in Acap1 Possesses Key Features of the Bar Domain in Promoting Membrane Curvature.
Dev.Cell, 31, 2014
|
|
4CKG
 
 | | Helical reconstruction of ACAP1(BAR-PH domain) decorated membrane tubules by cryo-electron microscopy | | Descriptor: | ARF-GAP WITH COILED-COIL, ANK REPEAT AND PH DOMAIN-CONTAINING PROTEIN 1 | | Authors: | Pang, X.Y, Fan, J, Zhang, Y, Zhang, K, Gao, B.Q, Ma, J, Li, J, Deng, Y.C, Zhou, Q.J, Hsu, V, Sun, F. | | Deposit date: | 2014-01-06 | | Release date: | 2014-10-15 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (15 Å) | | Cite: | A Ph Domain in Acap1 Possesses Key Features of the Bar Domain in Promoting Membrane Curvature.
Dev.Cell, 31, 2014
|
|
1M5Z
 
 | | The PDZ7 of Glutamate Receptor Interacting Protein Binds to its Target via a Novel Hydrophobic Surface Area | | Descriptor: | AMPA receptor interacting protein | | Authors: | Feng, W, Fan, J, Jiang, M, Shi, Y, Zhang, M. | | Deposit date: | 2002-07-11 | | Release date: | 2002-11-06 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | The PDZ7 of Glutamate Receptor Interacting Protein Binds to its Target via a Novel Hydrophobic Surface Area
J.Biol.Chem., 277, 2002
|
|
1RSO
 
 | | Hetero-tetrameric L27 (Lin-2, Lin-7) domain complexes as organization platforms of supra-molecular assemblies | | Descriptor: | Peripheral plasma membrane protein CASK, Presynaptic protein SAP97 | | Authors: | Feng, W, Long, J.-F, Fan, J.-S, Suetake, T, Zhang, M. | | Deposit date: | 2003-12-09 | | Release date: | 2004-05-04 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | The tetrameric L27 domain complex as an organization platform for supramolecular assemblies
NAT.STRUCT.MOL.BIOL., 11, 2004
|
|
6M5C
 
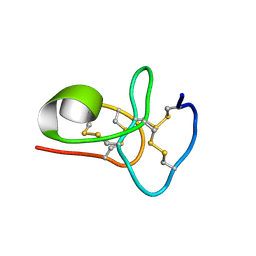 | | Solution structure of avenatide aV1 | | Descriptor: | avenatide aV1 | | Authors: | Tay, S.V, Wong, K.H, Huang, J.Y, Fan, J.S, Yang, D.W, Tam, J.P. | | Deposit date: | 2020-03-10 | | Release date: | 2021-03-10 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Solution structure of avenatide aV1
To Be Published
|
|
7YAV
 
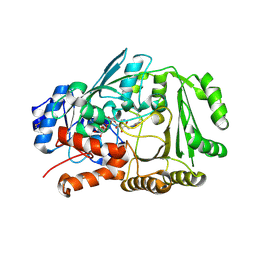 | | Crystal structure of Diels-Alderase MaDA1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, FLAVIN-ADENINE DINUCLEOTIDE, MaDA1 | | Authors: | Lei, X.G, Chen, R.C, Du, X.X, Yang, J, Fan, J.P, Guo, N.X, Ding, Q. | | Deposit date: | 2022-06-28 | | Release date: | 2024-01-24 | | Last modified: | 2024-09-11 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The evolutionary origin of naturally occurring intermolecular Diels-Alderases from Morus alba.
Nat Commun, 15, 2024
|
|
7YP0
 
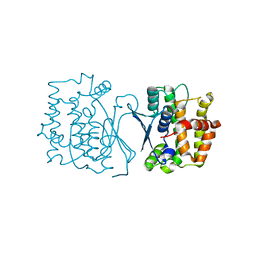 | | Crystal structure of CtGST | | Descriptor: | Glutathione S-transferase | | Authors: | Yang, J, Fan, J.P, Lei, X.G. | | Deposit date: | 2022-08-02 | | Release date: | 2024-02-07 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Enzymatic Degradation of Deoxynivalenol with the Engineered Detoxification Enzyme Fhb7.
Jacs Au, 4, 2024
|
|
5JQM
 
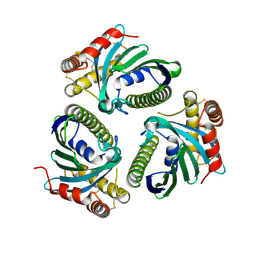 | | Crystal Structure of Phosphatidic acid Transporter Ups1/Mdm35 Void of Bound Phospholipid from Saccharomyces Cerevisiae at 1.5 Angstroms Resolution | | Descriptor: | Protein UPS1, mitochondrial,Mitochondrial distribution and morphology protein 35 | | Authors: | Lu, J, Chan, K.C, Zhai, Y, Fan, J, Sun, F. | | Deposit date: | 2016-05-05 | | Release date: | 2017-07-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Molecular mechanism of mitochondrial phosphatidate transfer by Ups1
Commun Biol, 2020
|
|
5JQL
 
 | | Crystal Structure of Phosphatidic acid Transporter Ups1/Mdm35 Void of Bound Phospholipid from Saccharomyces Cerevisiae at 2.9 Angstroms Resolution | | Descriptor: | Mitochondrial distribution and morphology protein 35, Protein UPS1, mitochondrial | | Authors: | Lu, J, Chan, K.C, Zhai, Y, Fan, J, Sun, F. | | Deposit date: | 2016-05-05 | | Release date: | 2017-07-12 | | Last modified: | 2020-09-02 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Molecular mechanism of mitochondrial phosphatidate transfer by Ups1
Commun Biol, 2020
|
|
8SQF
 
 | | OXA-48 bound to inhibitor CDD-2725 | | Descriptor: | (1M)-3'-(benzyloxy)-5-hydroxy[1,1'-biphenyl]-3,4'-dicarboxylic acid, BICARBONATE ION, Beta-lactamase | | Authors: | Park, S, Judge, A, Fan, J, Sankaran, B, Prasad, B.V.V, Palzkill, T. | | Deposit date: | 2023-05-04 | | Release date: | 2024-01-03 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Exploiting the Carboxylate-Binding Pocket of beta-Lactamase Enzymes Using a Focused DNA-Encoded Chemical Library.
J.Med.Chem., 67, 2024
|
|
8SQG
 
 | | OXA-48 bound to inhibitor CDD-2801 | | Descriptor: | (1M)-3'-(benzyloxy)-5-[2-(methylamino)-2-oxoethoxy][1,1'-biphenyl]-3,4'-dicarboxylic acid, BICARBONATE ION, Beta-lactamase | | Authors: | Park, S, Judge, A, Fan, J, Sankaran, B, Palzkill, T. | | Deposit date: | 2023-05-04 | | Release date: | 2024-01-03 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Exploiting the Carboxylate-Binding Pocket of beta-Lactamase Enzymes Using a Focused DNA-Encoded Chemical Library.
J.Med.Chem., 67, 2024
|
|
7DFE
 
 | | NMR structure of TuSp2-RP | | Descriptor: | B6 protein | | Authors: | Lin, Z, Fan, T, Fan, J. | | Deposit date: | 2020-11-07 | | Release date: | 2021-11-17 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | 1H, 15N and 13C resonance assignments of a repetitive domain of tubuliform spidroin 2
Biomol.Nmr Assign., 15, 2021
|
|
2M0M
 
 | | Structural Characterization of Minor Ampullate Spidroin Domains and their Distinct Roles in Fibroin Solubility and Fiber Formation | | Descriptor: | Minor ampullate fibroin 1 | | Authors: | Yang, D, Gao, Z, Lin, Z, Huang, W, Lai, C, Fan, J. | | Deposit date: | 2012-10-30 | | Release date: | 2013-03-27 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural characterization of minor ampullate spidroin domains and their distinct roles in fibroin solubility and fiber formation
Plos One, 8, 2013
|
|
1PWJ
 
 | | Structure of the Monomeric 8-kDa Dynein Light Chain and Mechanism of Domain Swapped Dimer Assembly | | Descriptor: | dynein light chain-2 | | Authors: | Wang, W, Lo, K.W.-H, Kan, H.-M, Fan, J.-S, Zhang, M. | | Deposit date: | 2003-07-02 | | Release date: | 2003-10-21 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structure of the Monomeric 8-kDa Dynein Light Chain and Mechanism of the Domain-swapped Dimer Assembly
J.Biol.Chem., 278, 2003
|
|
1PWK
 
 | | Structure of the Monomeric 8-kDa Dynein Light Chain and Mechanism of Domain Swapped Dimer Assembly | | Descriptor: | dynein light chain-2 | | Authors: | Wang, W, Lo, K.W.-H, Kan, H.-M, Fan, J.-S, Zhang, M. | | Deposit date: | 2003-07-02 | | Release date: | 2003-10-21 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structure of the Monomeric 8-kDa Dynein Light Chain and Mechanism of the Domain-swapped Dimer Assembly
J.Biol.Chem., 278, 2003
|
|
5H3D
 
 | | Helical structure of membrane tubules decorated by ACAP1 (BARPH doamin) protein by cryo-electron microscopy and MD simulation | | Descriptor: | Arf-GAP with coiled-coil, ANK repeat and PH domain-containing protein 1 | | Authors: | Chan, C, Pang, X.Y, Zhang, Y, Sun, F, Fan, J. | | Deposit date: | 2016-10-22 | | Release date: | 2019-01-16 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (14 Å) | | Cite: | ACAP1 assembles into an unusual protein lattice for membrane deformation through multiple stages.
Plos Comput.Biol., 15, 2019
|
|
2K3Q
 
 | |
