6F2X
 
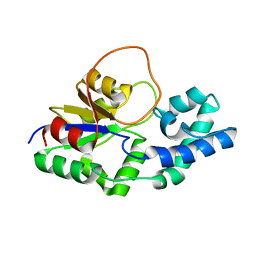 | | Structural characterization of the Mycobacterium tuberculosis Protein Tyrosine Kinase A (PtkA) | | Descriptor: | Protein Tyrosine Kinase A | | Authors: | Niesteruk, A, Jonker, H.R.A, Sreeramulu, S, Richter, C, Hutchison, M, Linhard, V, Schwalbe, H. | | Deposit date: | 2017-11-27 | | Release date: | 2018-07-04 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | The domain architecture of PtkA, the first tyrosine kinase fromMycobacterium tuberculosis, differs from the conventional kinase architecture.
J. Biol. Chem., 293, 2018
|
|
5IA4
 
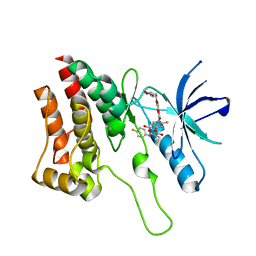 | | Crystal Structure of Ephrin A2 (EphA2) Receptor Protein Kinase with foretinib (XL880) | | Descriptor: | 1,2-ETHANEDIOL, Ephrin type-A receptor 2, N-(3-fluoro-4-{[6-methoxy-7-(3-morpholin-4-ylpropoxy)quinolin-4-yl]oxy}phenyl)-N'-(4-fluorophenyl)cyclopropane-1,1-dicarboxamide | | Authors: | Kudlinzki, D, Linhard, V.L, Gande, S.L, Sreeramulu, S, Saxena, K, Heinzlmeir, S, Medard, G, Kuester, B, Schwalbe, H. | | Deposit date: | 2016-02-21 | | Release date: | 2016-11-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.797 Å) | | Cite: | Chemical Proteomics and Structural Biology Define EPHA2 Inhibition by Clinical Kinase Drugs.
ACS Chem. Biol., 11, 2016
|
|
6FT2
 
 | |
7JOZ
 
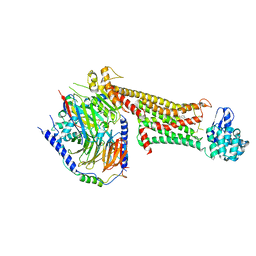 | | Crystal structure of dopamine D1 receptor in complex with G protein and a non-catechol agonist | | Descriptor: | 6-{4-[(furo[3,2-c]pyridin-4-yl)oxy]-2-methylphenyl}-1,5-dimethylpyrimidine-2,4(1H,3H)-dione, Endolysin,D(1A) dopamine receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Sun, B, Feng, D, Chu, M.L, Fish, I, Kelm, S, Lebon, F, Lovera, S, Valade, A, Wood, M, Ceska, T, Kobilka, T.S, Sands, Z, Kobilka, B.K. | | Deposit date: | 2020-08-07 | | Release date: | 2021-04-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Crystal structure of dopamine D1 receptor in complex with G protein and a non-catechol agonist.
Nat Commun, 12, 2021
|
|
6FLJ
 
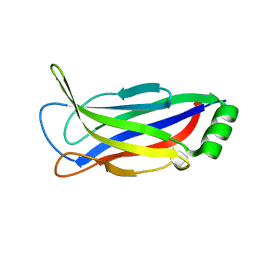 | |
6FLK
 
 | |
5HEI
 
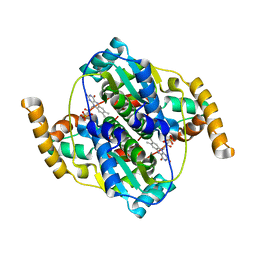 | | Structure of B. megaterium NfrA2 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, FLAVIN MONONUCLEOTIDE, NfrA2, ... | | Authors: | Vigouroux, A, Morera, S. | | Deposit date: | 2016-01-06 | | Release date: | 2016-04-06 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.84 Å) | | Cite: | Functional and structural characterization of two Bacillus megaterium nitroreductases biotransforming the herbicide mesotrione.
Biochem.J., 473, 2016
|
|
4Z8E
 
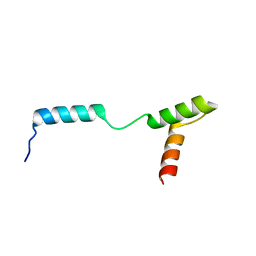 | | TEAD DBD mutant -deltaL1 | | Descriptor: | Transcriptional enhancer factor TEF-1 | | Authors: | Lee, D.-S, Albarado, D.C, Vonrhein, C, Raman, C.S, Veeraraghavan, S. | | Deposit date: | 2015-04-08 | | Release date: | 2016-04-13 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.092 Å) | | Cite: | A Potential Structural Switch for Regulating DNA-Binding by TEAD Transcription Factors.
J.Mol.Biol., 428, 2016
|
|
4ZE8
 
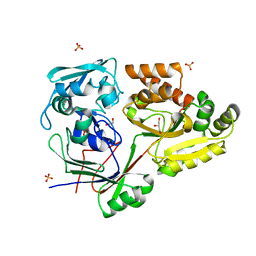 | | PBP AccA from A. tumefaciens C58 | | Descriptor: | 1,2-ETHANEDIOL, ABC transporter, substrate binding protein (Agrocinopines A and B), ... | | Authors: | El Sahili, A, Morera, S. | | Deposit date: | 2015-04-20 | | Release date: | 2015-08-19 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | A Pyranose-2-Phosphate Motif Is Responsible for Both Antibiotic Import and Quorum-Sensing Regulation in Agrobacterium tumefaciens.
Plos Pathog., 11, 2015
|
|
7SR2
 
 | | Crystal structure of the human SNX25 regulator of G-protein signalling (RGS) domain | | Descriptor: | ACETATE ION, LEUCINE, Sorting nexin-25, ... | | Authors: | Collins, B.M, Paul, B, Weeratunga, S. | | Deposit date: | 2021-11-07 | | Release date: | 2021-11-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Structural Predictions of the SNX-RGS Proteins Suggest They Belong to a New Class of Lipid Transfer Proteins.
Front Cell Dev Biol, 10, 2022
|
|
7SR1
 
 | |
4QVK
 
 | | Apo-crystal structure of Podospora anserina methyltransferase PaMTH1 | | Descriptor: | 1,2-ETHANEDIOL, PaMTH1 Methyltransferase | | Authors: | Kudlinzki, D, Linhard, V.L, Chatterjee, D, Saxena, K, Sreeramulu, S, Schwalbe, H. | | Deposit date: | 2014-07-15 | | Release date: | 2015-05-27 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Structure and Biophysical Characterization of the S-Adenosylmethionine-dependent O-Methyltransferase PaMTH1, a Putative Enzyme Accumulating during Senescence of Podospora anserina.
J.Biol.Chem., 290, 2015
|
|
3IWJ
 
 | | Crystal structure of aminoaldehyde dehydrogenase 2 from Pisum sativum (PsAMADH2) | | Descriptor: | GLYCEROL, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, Putative aminoaldehyde dehydrogenase, ... | | Authors: | Kopecny, D, Morera, S, Briozzo, P. | | Deposit date: | 2009-09-02 | | Release date: | 2010-01-19 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural and functional characterization of plant aminoaldehyde dehydrogenase from Pisum sativum with a broad specificity for natural and synthetic aminoaldehydes.
J.Mol.Biol., 396, 2010
|
|
3IWK
 
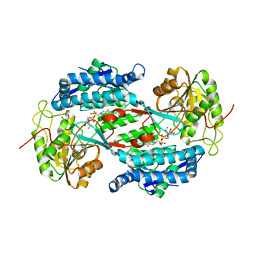 | | Crystal structure of aminoaldehyde dehydrogenase 1 from Pisum sativum (PsAMADH1) | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, Aminoaldehyde dehydrogenase, GLYCEROL, ... | | Authors: | Kopecny, D, Morera, S, Briozzo, P. | | Deposit date: | 2009-09-02 | | Release date: | 2010-01-19 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural and functional characterization of plant aminoaldehyde dehydrogenase from Pisum sativum with a broad specificity for natural and synthetic aminoaldehydes.
J.Mol.Biol., 396, 2010
|
|
1ANW
 
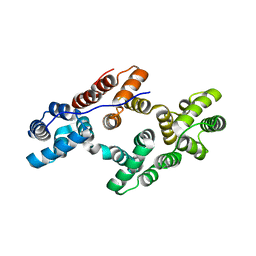 | | THE EFFECT OF METAL BINDING ON THE STRUCTURE OF ANNEXIN V AND IMPLICATIONS FOR MEMBRANE BINDING | | Descriptor: | ANNEXIN V, CALCIUM ION | | Authors: | Lewit-Bentley, A, Morera, S, Huber, R, Bodo, G. | | Deposit date: | 1993-10-26 | | Release date: | 1994-12-20 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The effect of metal binding on the structure of annexin V and implications for membrane binding.
Eur.J.Biochem., 210, 1992
|
|
5GY7
 
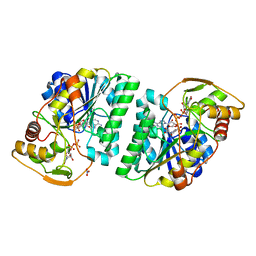 | | X-Ray structure of H243I mutant of UDP-Galactose 4-epimerase from E.coli:evidence for existence of open and closed active site during catalysis. | | Descriptor: | GLYCEROL, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, NITRATE ION, ... | | Authors: | Singh, N, Tiwari, P, Phulera, S, Dixit, A, Choudhury, D. | | Deposit date: | 2016-09-21 | | Release date: | 2016-11-30 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | X-Ray structure of H243I mutant of UDP-Galactose 4-epimerase from E.coli:evidence for existence of open and closed active site during catalysis.
To Be Published
|
|
4P0I
 
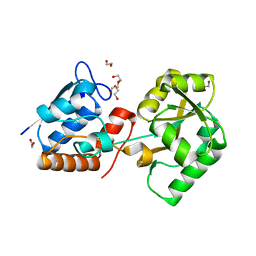 | | Structure of the PBP NocT | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, Nopaline-binding periplasmic protein | | Authors: | Vigouroux, A, Morera, S. | | Deposit date: | 2014-02-21 | | Release date: | 2014-10-22 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Agrobacterium uses a unique ligand-binding mode for trapping opines and acquiring a competitive advantage in the niche construction on plant host.
Plos Pathog., 10, 2014
|
|
4ZE9
 
 | | Se-PBP AccA from A. tumefaciens C58 in complex with agrocinopine A | | Descriptor: | 2-O-phosphono-alpha-L-arabinopyranose, ABC transporter substrate-binding protein, beta-D-fructofuranose-(2-1)-alpha-D-glucopyranose | | Authors: | El Sahili, A, Guimaraes, B.G, Morera, S. | | Deposit date: | 2015-04-20 | | Release date: | 2015-08-19 | | Last modified: | 2023-06-14 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | A Pyranose-2-Phosphate Motif Is Responsible for Both Antibiotic Import and Quorum-Sensing Regulation in Agrobacterium tumefaciens.
Plos Pathog., 11, 2015
|
|
4ZED
 
 | |
4ZEI
 
 | |
4ZEC
 
 | | PBP AccA from A. tumefaciens C58 in complex with agrocin 84 | | Descriptor: | 1,2-ETHANEDIOL, ABC transporter, substrate binding protein (Agrocinopines A and B), ... | | Authors: | El Sahili, A, Morera, S. | | Deposit date: | 2015-04-20 | | Release date: | 2015-08-19 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | A Pyranose-2-Phosphate Motif Is Responsible for Both Antibiotic Import and Quorum-Sensing Regulation in Agrobacterium tumefaciens.
Plos Pathog., 11, 2015
|
|
7JN3
 
 | | Cryo-EM structure of Rous sarcoma virus cleaved synaptic complex (CSC) with HIV-1 integrase strand transfer inhibitor MK-2048 | | Descriptor: | (6S)-2-(3-chloro-4-fluorobenzyl)-8-ethyl-10-hydroxy-N,6-dimethyl-1,9-dioxo-1,2,6,7,8,9-hexahydropyrazino[1',2':1,5]pyrrolo[2,3-d]pyridazine-4-carboxamide, DNA (5'-D(*AP*AP*TP*GP*TP*TP*GP*TP*CP*TP*TP*AP*TP*GP*CP*AP*AP*T)-3'), DNA (5'-D(*AP*TP*TP*GP*CP*AP*TP*AP*AP*GP*AP*CP*AP*AP*CP*A)-3'), ... | | Authors: | Pandey, K.K, Bera, S, Shi, K, Aihara, H, Grandgenett, D.P. | | Deposit date: | 2020-08-03 | | Release date: | 2021-03-17 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.21 Å) | | Cite: | Cryo-EM structure of the Rous sarcoma virus octameric cleaved synaptic complex intasome.
Commun Biol, 4, 2021
|
|
7KUI
 
 | | Cryo-EM structure of Rous sarcoma virus cleaved synaptic complex (CSC) with HIV-1 integrase strand transfer inhibitor MK-2048. CIC region of a cluster identified by 3-dimensional variability analysis in cryoSPARC. | | Descriptor: | (6S)-2-(3-chloro-4-fluorobenzyl)-8-ethyl-10-hydroxy-N,6-dimethyl-1,9-dioxo-1,2,6,7,8,9-hexahydropyrazino[1',2':1,5]pyrrolo[2,3-d]pyridazine-4-carboxamide, DNA (5'-D(*AP*AP*TP*GP*TP*TP*GP*TP*CP*TP*TP*AP*TP*GP*CP*AP*AP*T)-3'), DNA (5'-D(*AP*TP*TP*GP*CP*AP*TP*AP*AP*GP*AP*CP*AP*AP*CP*A)-3'), ... | | Authors: | Pandey, K.K, Bera, S, Shi, K, Aihara, H, Grandgenett, D.P. | | Deposit date: | 2020-11-25 | | Release date: | 2021-03-17 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Cryo-EM structure of the Rous sarcoma virus octameric cleaved synaptic complex intasome.
Commun Biol, 4, 2021
|
|
7KU7
 
 | | Cryo-EM structure of Rous sarcoma virus cleaved synaptic complex (CSC) with HIV-1 integrase strand transfer inhibitor MK-2048. Cluster identified by 3-dimensional variability analysis in cryoSPARC. | | Descriptor: | (6S)-2-(3-chloro-4-fluorobenzyl)-8-ethyl-10-hydroxy-N,6-dimethyl-1,9-dioxo-1,2,6,7,8,9-hexahydropyrazino[1',2':1,5]pyrrolo[2,3-d]pyridazine-4-carboxamide, DNA (5'-D(*AP*AP*TP*GP*TP*TP*GP*TP*CP*TP*TP*AP*TP*GP*CP*AP*AP*T)-3'), DNA (5'-D(*AP*TP*TP*GP*CP*AP*TP*AP*AP*GP*AP*CP*AP*AP*CP*A)-3'), ... | | Authors: | Pandey, K.K, Bera, S, Shi, K, Aihara, H, Grandgenett, D.P. | | Deposit date: | 2020-11-24 | | Release date: | 2021-03-17 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Cryo-EM structure of the Rous sarcoma virus octameric cleaved synaptic complex intasome.
Commun Biol, 4, 2021
|
|
4ZA6
 
 | |
