5G5L
 
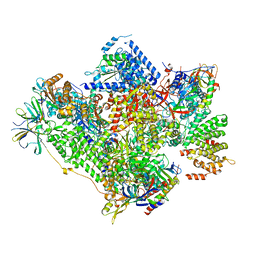 | | RNA polymerase I-Rrn3 complex at 4.8 A resolution | | Descriptor: | DNA-DIRECTED RNA POLYMERASE I SUBUNIT RPA12, DNA-DIRECTED RNA POLYMERASE I SUBUNIT RPA135, DNA-DIRECTED RNA POLYMERASE I SUBUNIT RPA14, ... | | Authors: | Engel, C, Plitzko, J, Cramer, P. | | Deposit date: | 2016-05-26 | | Release date: | 2016-07-27 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | RNA Polymerase I-Rrn3 Complex at 4.8 A Resolution
Nat.Commun., 7, 2016
|
|
1XUD
 
 | | Matrix metalloproteinase-13 complexed with non-zinc binding inhibitor | | Descriptor: | CALCIUM ION, Collagenase 3, N,N'-BIS(4-FLUORO-3-METHYLBENZYL)PYRIMIDINE-4,6-DICARBOXAMIDE, ... | | Authors: | Engel, C.K, Wendt, K.U. | | Deposit date: | 2004-10-26 | | Release date: | 2005-10-26 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis for the highly selective inhibition of MMP-13.
Chem.Biol., 12, 2005
|
|
1XUR
 
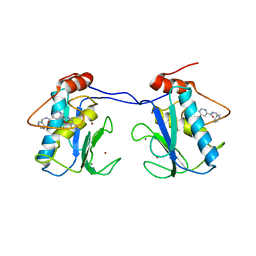 | | Matrix metalloproteinase-13 complexed with non-zinc binding inhibitor | | Descriptor: | CALCIUM ION, Collagenase 3, N,N'-BIS(PYRIDIN-3-YLMETHYL)PYRIMIDINE-4,6-DICARBOXAMIDE, ... | | Authors: | Engel, C.K, Wendt, K.U. | | Deposit date: | 2004-10-26 | | Release date: | 2005-10-26 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural basis for the highly selective inhibition of MMP-13.
Chem.Biol., 12, 2005
|
|
2DUB
 
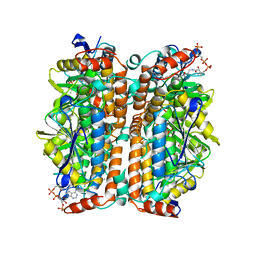 | | ENOYL-COA HYDRATASE COMPLEXED WITH OCTANOYL-COA | | Descriptor: | 2-ENOYL-COA HYDRATASE, OCTANOYL-COENZYME A | | Authors: | Engel, C.K, Wierenga, R.K. | | Deposit date: | 1997-04-28 | | Release date: | 1998-04-29 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The crystal structure of enoyl-CoA hydratase complexed with octanoyl-CoA reveals the structural adaptations required for binding of a long chain fatty acid-CoA molecule.
J.Mol.Biol., 275, 1998
|
|
1XUC
 
 | | Matrix metalloproteinase-13 complexed with non-zinc binding inhibitor | | Descriptor: | CALCIUM ION, Collagenase 3, N,N'-BIS(3-METHYLBENZYL)PYRIMIDINE-4,6-DICARBOXAMIDE, ... | | Authors: | Engel, C.K, Wendt, K.U. | | Deposit date: | 2004-10-26 | | Release date: | 2005-10-26 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural basis for the highly selective inhibition of MMP-13
Chem.Biol., 12, 2005
|
|
5O7X
 
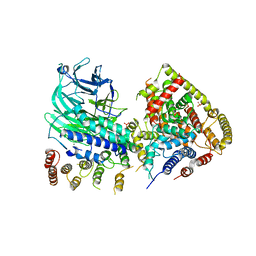 | | CRYSTAL STRUCTURE OF S. CEREVISIAE CORE FACTOR AT 3.2A RESOLUTION | | Descriptor: | MAGNESIUM ION, RNA polymerase I-specific transcription initiation factor RRN11, RNA polymerase I-specific transcription initiation factor RRN6, ... | | Authors: | Engel, C, Gubbey, T, Neyer, S, Sainsbury, S, Oberthuer, C, Baejen, C, Bernecky, C, Cramer, P. | | Deposit date: | 2017-06-09 | | Release date: | 2017-08-02 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural Basis of RNA Polymerase I Transcription Initiation.
Cell, 169, 2017
|
|
4C2M
 
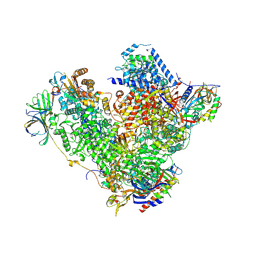 | | Structure of RNA polymerase I at 2.8 A resolution | | Descriptor: | DNA-DIRECTED RNA POLYMERASE I SUBUNIT RPA12, DNA-DIRECTED RNA POLYMERASE I SUBUNIT RPA135, DNA-DIRECTED RNA POLYMERASE I SUBUNIT RPA14, ... | | Authors: | Engel, C, Sainsbury, S, Cheung, A.C, Kostrewa, D, Cramer, P. | | Deposit date: | 2013-08-19 | | Release date: | 2013-10-23 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | RNA Polymerase I Structure and Transcription Regulation.
Nature, 502, 2013
|
|
5N61
 
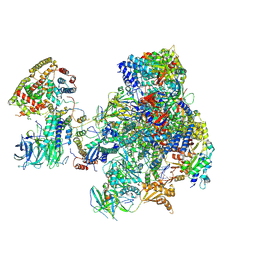 | | RNA polymerase I initially transcribing complex | | Descriptor: | DNA-directed RNA polymerase I subunit RPA12, DNA-directed RNA polymerase I subunit RPA135, DNA-directed RNA polymerase I subunit RPA14, ... | | Authors: | Engel, C, Gubbey, T, Neyer, S, Sainsbury, S, Oberthuer, C, Baejen, C, Bernecky, C, Cramer, P. | | Deposit date: | 2017-02-14 | | Release date: | 2017-04-05 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural Basis of RNA Polymerase I Transcription Initiation.
Cell, 169, 2017
|
|
5N5Z
 
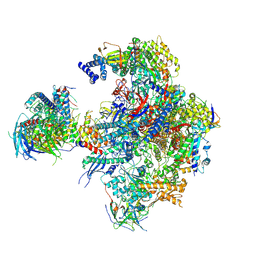 | | Cryo-EM structure of RNA polymerase I in complex with Rrn3 and Core Factor (Orientation II) | | Descriptor: | DNA-directed RNA polymerase I subunit RPA12, DNA-directed RNA polymerase I subunit RPA135, DNA-directed RNA polymerase I subunit RPA14, ... | | Authors: | Engel, C, Gubbey, T, Neyer, S, Sainsbury, S, Oberthuer, C, Baejen, C, Bernecky, C, Cramer, P. | | Deposit date: | 2017-02-14 | | Release date: | 2017-04-05 | | Last modified: | 2018-10-03 | | Method: | ELECTRON MICROSCOPY (7.7 Å) | | Cite: | Structural Basis of RNA Polymerase I Transcription Initiation.
Cell, 169, 2017
|
|
5N60
 
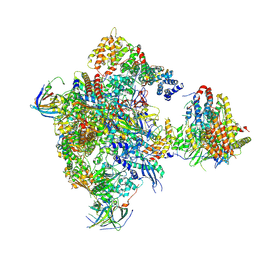 | | Cryo-EM structure of RNA polymerase I in complex with Rrn3 and Core Factor (Orientation I) | | Descriptor: | DNA-directed RNA polymerase I subunit RPA12, DNA-directed RNA polymerase I subunit RPA135, DNA-directed RNA polymerase I subunit RPA14, ... | | Authors: | Engel, C, Gubbey, T, Neyer, S, Sainsbury, S, Oberthuer, C, Baejen, C, Bernecky, C, Cramer, P. | | Deposit date: | 2017-02-14 | | Release date: | 2017-04-05 | | Last modified: | 2018-10-03 | | Method: | ELECTRON MICROSCOPY (7.7 Å) | | Cite: | Structural Basis of RNA Polymerase I Transcription Initiation.
Cell, 169, 2017
|
|
5N5Y
 
 | | Cryo-EM structure of RNA polymerase I in complex with Rrn3 and Core Factor (Orientation III) | | Descriptor: | DNA-directed RNA polymerase I subunit RPA12, DNA-directed RNA polymerase I subunit RPA135, DNA-directed RNA polymerase I subunit RPA14, ... | | Authors: | Engel, C, Gubbey, T, Neyer, S, Sainsbury, S, Oberthuer, C, Baejen, C, Bernecky, C, Cramer, P. | | Deposit date: | 2017-02-14 | | Release date: | 2017-04-05 | | Last modified: | 2018-10-03 | | Method: | ELECTRON MICROSCOPY (7.7 Å) | | Cite: | Structural Basis of RNA Polymerase I Transcription Initiation.
Cell, 169, 2017
|
|
6TPS
 
 | | early intermediate RNA Polymerase I Pre-initiation complex - eiPIC | | Descriptor: | DNA foreign, DNA-directed RNA polymerase I subunit RPA12, DNA-directed RNA polymerase I subunit RPA135, ... | | Authors: | Pilsl, M, Engel, C. | | Deposit date: | 2019-12-14 | | Release date: | 2020-03-18 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.54 Å) | | Cite: | Structural basis of RNA polymerase I pre-initiation complex formation and promoter melting.
Nat Commun, 11, 2020
|
|
8CRO
 
 | | Cryo-EM structure of Pyrococcus furiosus transcription elongation complex | | Descriptor: | DNA Non-Template Strand, DNA Template Strand, DNA-directed RNA polymerase subunit Rpo10, ... | | Authors: | Tarau, D.M, Grunberger, F, Reichelt, R, Heiss, F.B, Pilsl, M, Hausner, W, Engel, C, Grohmann, D. | | Deposit date: | 2023-03-08 | | Release date: | 2024-04-17 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural basis of archaeal RNA polymerase transcription elongation and Spt4/5 recruitment.
Nucleic Acids Res., 52, 2024
|
|
8A43
 
 | | Human RNA polymerase I | | Descriptor: | DNA-directed RNA polymerase I subunit RPA1, DNA-directed RNA polymerase I subunit RPA12, DNA-directed RNA polymerase I subunit RPA2, ... | | Authors: | Daiss, J.L, Pilsl, M, Straub, K, Bleckmann, A, Hoecherl, M, Heiss, F.B, Abascal-Palacios, G, Ramsay, E, Tluckova, K, Mars, J.C, Fuertges, T, Bruckmann, A, Rudack, T, Bernecky, C, Lamour, V, Panov, K, Vannini, A, Moss, T, Engel, C. | | Deposit date: | 2022-06-10 | | Release date: | 2022-08-31 | | Last modified: | 2022-11-02 | | Method: | ELECTRON MICROSCOPY (4.09 Å) | | Cite: | The human RNA polymerase I structure reveals an HMG-like docking domain specific to metazoans.
Life Sci Alliance, 5, 2022
|
|
4YM7
 
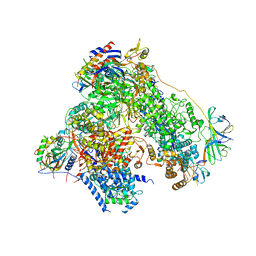 | | RNA polymerase I structure with an alternative dimer hinge | | Descriptor: | DNA-directed RNA polymerase I subunit RPA12, DNA-directed RNA polymerase I subunit RPA135, DNA-directed RNA polymerase I subunit RPA14, ... | | Authors: | Kostrewa, D, Kuhn, C.-D, Engel, C, Cramer, P. | | Deposit date: | 2015-03-06 | | Release date: | 2015-09-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (5.5 Å) | | Cite: | An alternative RNA polymerase I structure reveals a dimer hinge.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4HX5
 
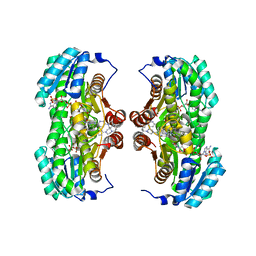 | | Crystal structure of 11 beta-HSD1 in complex with SAR184841 | | Descriptor: | 4-[5-(4-tert-butylpiperazin-1-yl)pyridin-2-yl]-N-[(1R,2S,3S,5S,7s)-5-carbamoyltricyclo[3.3.1.1~3,7~]dec-2-yl]-3,4-dihydroquinoxaline-1(2H)-carboxamide, Corticosteroid 11-beta-dehydrogenase isozyme 1, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Loenze, P, Schimanski-Breves, S, Engel, C.K. | | Deposit date: | 2012-11-09 | | Release date: | 2013-05-08 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Discovery of SAR184841, a potent and long-lasting inhibitor of 11beta-hydroxysteroid dehydrogenase type 1, active in a physiopathological animal model of T2D
Bioorg.Med.Chem.Lett., 23, 2013
|
|
8ORQ
 
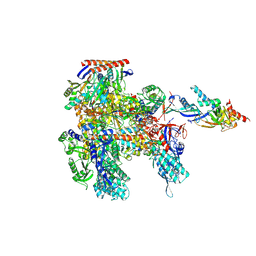 | | Cryo-EM structure of Pyrococcus furiosus apo form RNA polymerase open clamp conformation | | Descriptor: | DNA-directed RNA polymerase subunit Rpo10, DNA-directed RNA polymerase subunit Rpo11, DNA-directed RNA polymerase subunit Rpo12, ... | | Authors: | Tarau, D.M, Reichelt, R, Heiss, F.B, Pilsl, M, Hausner, W, Engel, C, Grohmann, D. | | Deposit date: | 2023-04-17 | | Release date: | 2024-04-24 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural basis of archaeal RNA polymerase transcription elongation and Spt4/5 recruitment.
Nucleic Acids Res., 52, 2024
|
|
8P2I
 
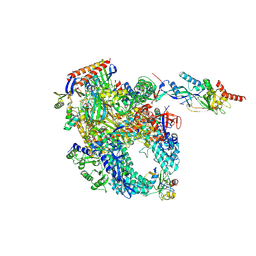 | | Cryo-EM structure of Pyrococcus furiosus apo form RNA polymerase contracted clamp conformation with Spt4/5 | | Descriptor: | DNA-directed RNA polymerase subunit Rpo10, DNA-directed RNA polymerase subunit Rpo11, DNA-directed RNA polymerase subunit Rpo12, ... | | Authors: | Tarau, D.M, Reichelt, R, Heiss, F.B, Pilsl, M, Hausner, W, Engel, C, Grohmann, D. | | Deposit date: | 2023-05-16 | | Release date: | 2024-04-24 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural basis of archaeal RNA polymerase transcription elongation and Spt4/5 recruitment.
Nucleic Acids Res., 52, 2024
|
|
8OKI
 
 | | Cryo-EM structure of Pyrococcus furiosus transcription elongation complex bound to Spt4/5 | | Descriptor: | DNA Non-Template Strand, DNA Template Strand, DNA-directed RNA polymerase subunit Rpo10, ... | | Authors: | Tarau, D.M, Reichelt, R, Heiss, F.B, Pilsl, M, Hausner, W, Engel, C, Grohmann, D. | | Deposit date: | 2023-03-28 | | Release date: | 2024-04-24 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.45 Å) | | Cite: | Structural basis of archaeal RNA polymerase transcription elongation and Spt4/5 recruitment.
Nucleic Acids Res., 52, 2024
|
|
7PZC
 
 | | Cryo-EM structure of the NLRP3 decamer bound to the inhibitor CRID3 | | Descriptor: | 1-(1,2,3,5,6,7-hexahydro-s-indacen-4-yl)-3-[4-(2-oxidanylpropan-2-yl)furan-2-yl]sulfonyl-urea, ADENOSINE-5'-DIPHOSPHATE, NACHT, ... | | Authors: | Hochheiser, I.V, Pilsl, M, Hagelueken, G, Engel, C, Geyer, M. | | Deposit date: | 2021-10-12 | | Release date: | 2022-01-26 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structure of the NLRP3 decamer bound to the cytokine release inhibitor CRID3.
Nature, 604, 2022
|
|
7AOC
 
 | | Schizosaccharomyces pombe RNA polymerase I (monomer) | | Descriptor: | DNA-directed RNA polymerase I subunit RPA12, DNA-directed RNA polymerase I subunit rpa1, DNA-directed RNA polymerase I subunit rpa14, ... | | Authors: | Heiss, F, Daiss, J, Becker, P, Engel, C. | | Deposit date: | 2020-10-14 | | Release date: | 2021-02-24 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.84 Å) | | Cite: | Conserved strategies of RNA polymerase I hibernation and activation.
Nat Commun, 12, 2021
|
|
7AOE
 
 | | Schizosaccharomyces pombe RNA polymerase I (elongation complex) | | Descriptor: | DNA-directed RNA polymerase I subunit RPA12, DNA-directed RNA polymerase I subunit rpa1, DNA-directed RNA polymerase I subunit rpa14, ... | | Authors: | Heiss, F, Daiss, J, Becker, P, Engel, C. | | Deposit date: | 2020-10-14 | | Release date: | 2021-02-24 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Conserved strategies of RNA polymerase I hibernation and activation.
Nat Commun, 12, 2021
|
|
8RBO
 
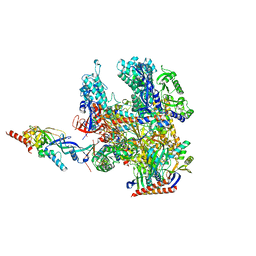 | | Cryo-EM structure of Pyrococcus furiosus apo form RNA polymerase contracted clamp conformation | | Descriptor: | DNA-directed RNA polymerase subunit Rpo10, DNA-directed RNA polymerase subunit Rpo11, DNA-directed RNA polymerase subunit Rpo12, ... | | Authors: | Tarau, D.M, Reichelt, R, Heiss, F.B, Pilsl, M, Hausner, W, Engel, C, Grohmann, D. | | Deposit date: | 2023-12-04 | | Release date: | 2024-04-24 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.02 Å) | | Cite: | Structural basis of archaeal RNA polymerase transcription elongation and Spt4/5 recruitment.
Nucleic Acids Res., 52, 2024
|
|
7AOD
 
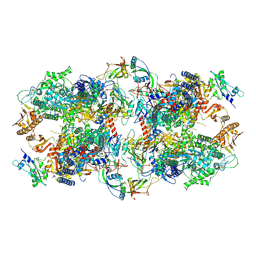 | | Schizosaccharomyces pombe RNA polymerase I (dimer) | | Descriptor: | DNA-directed RNA polymerase I subunit RPA12, DNA-directed RNA polymerase I subunit rpa1, DNA-directed RNA polymerase I subunit rpa14, ... | | Authors: | Heiss, F, Daiss, J, Becker, P, Engel, C. | | Deposit date: | 2020-10-14 | | Release date: | 2021-02-24 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Conserved strategies of RNA polymerase I hibernation and activation.
Nat Commun, 12, 2021
|
|
7AST
 
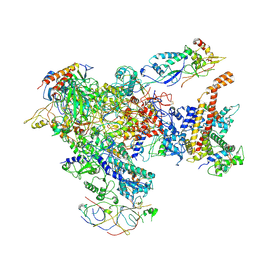 | | Apo Human RNA Polymerase III | | Descriptor: | DNA-directed RNA polymerase III subunit RPC1, DNA-directed RNA polymerase III subunit RPC10, DNA-directed RNA polymerase III subunit RPC2, ... | | Authors: | Ramsay, E.P, Abascal-Palacios, G, Daiss, J.L, King, H, Gouge, J, Pilsl, M, Beuron, F, Morris, E, Gunkel, P, Engel, C, Vannini, A. | | Deposit date: | 2020-10-28 | | Release date: | 2020-12-23 | | Last modified: | 2020-12-30 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structure of human RNA polymerase III.
Nat Commun, 11, 2020
|
|
