1EN5
 
 | | CRYSTAL STRUCTURE ANALYSIS OF THE E. COLI MANGANESE SUPEROXIDE DISMUTASE Y34F MUTANT | | Descriptor: | MANGANESE (II) ION, MANGANESE SUPEROXIDE DISMUTASE | | Authors: | Edwards, R.A, Whittaker, M.M, Baker, E.N, Whittaker, J.W, Jameson, G.B. | | Deposit date: | 2000-03-20 | | Release date: | 2001-06-27 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Outer sphere mutations perturb metal reactivity in manganese superoxide dismutase.
Biochemistry, 40, 2001
|
|
1EN4
 
 | | CRYSTAL STRUCTURE ANALYSIS OF THE E. COLI MANGANESE SUPEROXIDE DISMUTASE Q146H MUTANT | | Descriptor: | MANGANESE (II) ION, MANGANESE SUPEROXIDE DISMUTASE | | Authors: | Edwards, R.A, Whittaker, M.M, Baker, E.N, Whittaker, J.W, Jameson, G.B. | | Deposit date: | 2000-03-20 | | Release date: | 2001-06-27 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Outer sphere mutations perturb metal reactivity in manganese superoxide dismutase.
Biochemistry, 40, 2001
|
|
1EN6
 
 | | CRYSTAL STRUCTURE ANALYSIS OF THE E. COLI MANGANESE SUPEROXIDE DISMUTASE Q146L MUTANT | | Descriptor: | MANGANESE (II) ION, MANGANESE SUPEROXIDE DISMUTASE | | Authors: | Edwards, R.A, Whittaker, M.M, Baker, E.N, Whittaker, J.W, Jameson, G.B. | | Deposit date: | 2000-03-20 | | Release date: | 2001-06-27 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Outer sphere mutations perturb metal reactivity in manganese superoxide dismutase.
Biochemistry, 40, 2001
|
|
1I08
 
 | | CRYSTAL STRUCTURE ANALYSIS OF THE H30A MUTANT OF MANGANESE SUPEROXIDE DISMUTASE FROM E. COLI | | Descriptor: | MANGANESE (II) ION, MANGANESE SUPEROXIDE DISMUTASE | | Authors: | Edwards, R.A, Whittaker, M.M, Whittaker, J.W, Baker, E.N, Jameson, G.B. | | Deposit date: | 2001-01-29 | | Release date: | 2001-02-28 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Removing a hydrogen bond in the dimer interface of Escherichia coli manganese superoxide dismutase alters structure and reactivity.
Biochemistry, 40, 2001
|
|
1I0H
 
 | | CRYSTAL STRUCTURE OF THE E. COLI MANGANESE SUPEROXIDE DISMUTASE MUTANT Y174F AT 1.35 ANGSTROMS RESOLUTION. | | Descriptor: | MANGANESE (II) ION, MANGANESE SUPEROXIDE DISMUTASE Y174F MUTANT | | Authors: | Edwards, R.A, Whittaker, M.M, Whittaker, J.W, Baker, E.N, Jameson, G.B. | | Deposit date: | 2001-01-29 | | Release date: | 2001-02-28 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Removing a hydrogen bond in the dimer interface of Escherichia coli manganese superoxide dismutase alters structure and reactivity.
Biochemistry, 40, 2001
|
|
1MMM
 
 | | DISTINCT METAL ENVIRONMENT IN IRON-SUBSTITUTED MANGANESE SUPEROXIDE DISMUTASE PROVIDES A STRUCTURAL BASIS OF METAL SPECIFICITY | | Descriptor: | FE (III) ION, HYDROXIDE ION, PROTEIN (IRON-SUBSTITUTED MANGANESE SUPEROXIDE DISMUTASE) | | Authors: | Edwards, R.A, Whittaker, M.M, Whittaker, J.W, Jameson, G.B, Baker, E.N. | | Deposit date: | 1998-08-26 | | Release date: | 1998-09-02 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Distinct Metal Environment in Fe-Substituted Manganese Superoxide Dismutase Provides a Structural Basis of Metal Specificity
J.Am.Chem.Soc., 120, 1998
|
|
1VEW
 
 | | MANGANESE SUPEROXIDE DISMUTASE FROM ESCHERICHIA COLI | | Descriptor: | HYDROXIDE ION, MANGANESE (II) ION, MANGANESE SUPEROXIDE DISMUTASE | | Authors: | Edwards, R.A, Baker, H.M, Whittaker, M.M, Whittaker, J.W, Jameson, G.B, Baker, E.N. | | Deposit date: | 1998-01-20 | | Release date: | 1998-05-27 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of Escherichia coli manganese superoxide dismutase at 2.1-angstrom resolution.
J.Biol.Inorg.Chem., 3, 1998
|
|
9BIV
 
 | |
3D8A
 
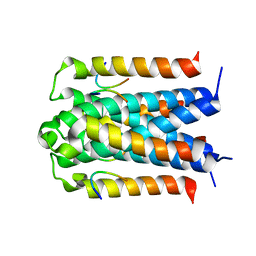 | | Co-crystal structure of TraM-TraD complex. | | Descriptor: | Protein traD, Relaxosome protein TraM | | Authors: | Glover, J.N.M, Lu, J, Wong, J.J, Edwards, R.A. | | Deposit date: | 2008-05-22 | | Release date: | 2008-09-09 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structural basis of specific TraD-TraM recognition during F plasmid-mediated bacterial conjugation.
Mol.Microbiol., 70, 2008
|
|
8UK7
 
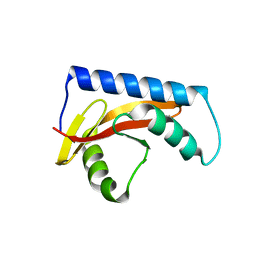 | |
2G9E
 
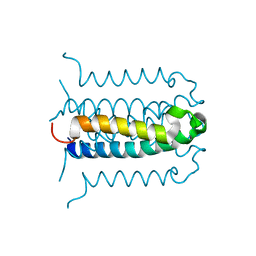 | | Protonation-mediated structural flexibility in the F conjugation regulatory protein, TRAM | | Descriptor: | Protein traM | | Authors: | Lu, J, Edwards, R.A, Wong, J.J, Manchak, J, Scott, P.G, Frost, L.S, Glover, J.N. | | Deposit date: | 2006-03-06 | | Release date: | 2006-06-13 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Protonation-mediated structural flexibility in the F conjugation regulatory protein, TraM.
Embo J., 25, 2006
|
|
7RGS
 
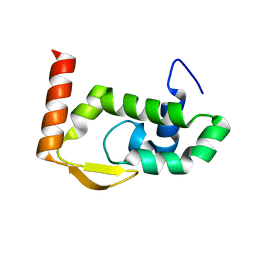 | | The crystal structure of RocC, containing FinO domain, 24-126 | | Descriptor: | Repressor of competence, RNA Chaperone | | Authors: | Kim, H.J, Edwards, R.A, Glover, J.N.M. | | Deposit date: | 2021-07-15 | | Release date: | 2022-11-09 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis for recognition of transcriptional terminator structures by ProQ/FinO domain RNA chaperones.
Nat Commun, 13, 2022
|
|
7RGT
 
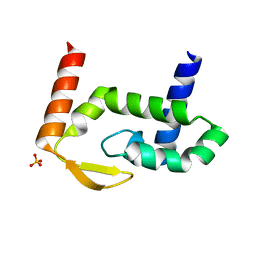 | | The crystal structure of RocC, containing FinO domain, 1-126 | | Descriptor: | Repressor of competence, RNA Chaperone, SULFATE ION | | Authors: | Kim, H.J, Edwards, R.A, Glover, J.N.M. | | Deposit date: | 2021-07-15 | | Release date: | 2022-11-09 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Structural basis for recognition of transcriptional terminator structures by ProQ/FinO domain RNA chaperones.
Nat Commun, 13, 2022
|
|
7RGU
 
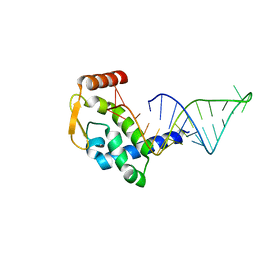 | |
4QPQ
 
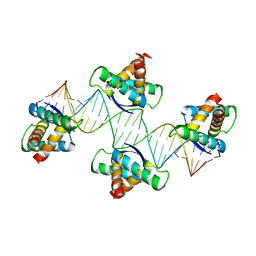 | | Mechanistic basis of plasmid-specific DNA binding of the F plasmid regulatory protein, TraM | | Descriptor: | Relaxosome protein TraM, sbmA DNA1, sbmA DNA2 | | Authors: | Peng, Y, Lu, J, Wong, J, Edwards, R.A, Frost, L.S, Glover, J.N.M. | | Deposit date: | 2014-06-24 | | Release date: | 2014-09-24 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.106 Å) | | Cite: | Mechanistic Basis of Plasmid-Specific DNA Binding of the F Plasmid Regulatory Protein, TraM.
J.Mol.Biol., 426, 2014
|
|
4QPO
 
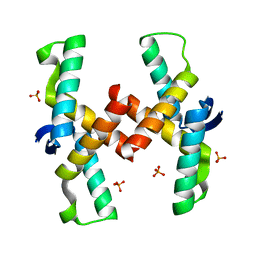 | | Mechanistic basis of plasmid-specific DNA binding of the F plasmid regulatory protein, TraM | | Descriptor: | PHOSPHATE ION, Relaxosome protein TraM | | Authors: | Peng, Y, Lu, J, Wong, J, Edwards, R.A, Frost, L.S, Glover, J.N.M. | | Deposit date: | 2014-06-24 | | Release date: | 2014-09-03 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.999 Å) | | Cite: | Mechanistic Basis of Plasmid-Specific DNA Binding of the F Plasmid Regulatory Protein, TraM.
J.Mol.Biol., 426, 2014
|
|
4WHV
 
 | | E3 ubiquitin-protein ligase RNF8 in complex with Ubiquitin-conjugating enzyme E2 N and Polyubiquitin-B | | Descriptor: | E3 ubiquitin-protein ligase RNF8, Polyubiquitin-B, Ubiquitin-conjugating enzyme E2 N, ... | | Authors: | Hodge, C.D, Edwards, R.A, Glover, J.N.M. | | Deposit date: | 2014-09-23 | | Release date: | 2015-09-30 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (8.3 Å) | | Cite: | RNF8 E3 Ubiquitin Ligase Stimulates Ubc13 E2 Conjugating Activity That Is Essential for DNA Double Strand Break Signaling and BRCA1 Tumor Suppressor Recruitment.
J.Biol.Chem., 291, 2016
|
|
1IXB
 
 | | CRYSTAL STRUCTURE OF THE E. COLI MANGANESE(II) SUPEROXIDE DISMUTASE MUTANT Y174F AT 0.90 ANGSTROMS RESOLUTION. | | Descriptor: | MANGANESE ION, 1 HYDROXYL COORDINATED, SUPEROXIDE DISMUTASE | | Authors: | Anderson, B.F, Edwards, R.A, Whittaker, M.M, Whittaker, J.W, Baker, E.N, Jameson, G.B. | | Deposit date: | 2002-06-18 | | Release date: | 2002-12-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (0.9 Å) | | Cite: | Structures at 0.90 A resolution of the oxidised and reduced forms of the Y174F mutant of the manganese superoxide dismutase from Escherichia coli
To be Published
|
|
1IX9
 
 | | Crystal Structure of the E. coli Manganase(III) superoxide dismutase mutant Y174F at 0.90 angstroms resolution. | | Descriptor: | MANGANESE (II) ION, Superoxide Dismutase | | Authors: | Anderson, B.F, Edwards, R.A, Whittaker, M.M, Whittaker, J.W, Baker, E.N, Jameson, G.B. | | Deposit date: | 2002-06-17 | | Release date: | 2002-12-17 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (0.9 Å) | | Cite: | Structures at 0.90 A resolution of the oxidised and reduced forms of the Y174F mutant of the manganese superoxide dismutase from Escherichia coli
To be Published
|
|
8D34
 
 | |
2ADO
 
 | | Crystal Structure Of The Brct Repeat Region From The Mediator of DNA damage checkpoint protein 1, MDC1 | | Descriptor: | Mediator of DNA damage checkpoint protein 1 | | Authors: | Lee, M.S, Edwards, R.A, Thede, G.L, Glover, J.N. | | Deposit date: | 2005-07-20 | | Release date: | 2005-08-02 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structure of the BRCT Repeat Domain of MDC1 and Its Specificity for the Free COOH-terminal End of the {gamma}-H2AX Histone Tail.
J.Biol.Chem., 280, 2005
|
|
3K0H
 
 | | The crystal structure of BRCA1 BRCT in complex with a minimal recognition tetrapeptide with an amidated C-terminus | | Descriptor: | Breast cancer type 1 susceptibility protein, CHLORIDE ION, NICKEL (II) ION, ... | | Authors: | Campbell, S.J, Edwards, R.A, Glover, J.N. | | Deposit date: | 2009-09-24 | | Release date: | 2010-03-02 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Comparison of the Structures and Peptide Binding Specificities of the BRCT Domains of MDC1 and BRCA1
Structure, 18, 2010
|
|
3K15
 
 | | Crystal Structure of BRCA1 BRCT D1840T in complex with a minimal recognition tetrapeptide with an amidated C-terminus | | Descriptor: | Breast cancer type 1 susceptibility protein, CHLORIDE ION, NICKEL (II) ION, ... | | Authors: | Campbell, S.J, Edwards, R.A, Glover, J.N. | | Deposit date: | 2009-09-25 | | Release date: | 2010-03-02 | | Last modified: | 2021-10-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Comparison of the Structures and Peptide Binding Specificities of the BRCT Domains of MDC1 and BRCA1
Structure, 18, 2010
|
|
3K16
 
 | | Crystal Structure of BRCA1 BRCT D1840T in complex with a minimal recognition tetrapeptide with a free carboxy C-terminus | | Descriptor: | Breast cancer type 1 susceptibility protein, CHLORIDE ION, NICKEL (II) ION, ... | | Authors: | Campbell, S.J, Edwards, R.A, Glover, J.N. | | Deposit date: | 2009-09-25 | | Release date: | 2010-03-02 | | Last modified: | 2021-10-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Comparison of the Structures and Peptide Binding Specificities of the BRCT Domains of MDC1 and BRCA1
Structure, 18, 2010
|
|
3K05
 
 | |
